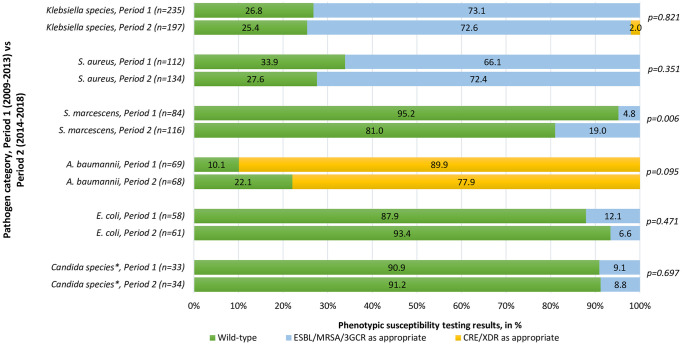
Fig 2. Phenotypic susceptibility patterns for the most commonly isolated pathogens in neonatal hospital-acquired bloodstream infections, Period 1 (2009–2013) vs Period 2 (2014–2018).
*Phenotypic susceptibility patterns established using the relevant year’s Clinical and Laboratory Standards Institute (CLSI) breakpoints. Isolates classified according to standard definitions for wild-type and acquired resistance, and reported as a proportion of the total number of isolates per species. Two-sided p-values were used. ESBL: extended-spectrum β-lactamase, detected by third-generation cephalosporin resistance; MRSA: methicillin-resistant S. aureus, detected by cefoxitin resistance;3GCR: 3rd-generation cephalosporin resistance in chromosomal ampC beta-lactamase producers; CRE: carbapenem-resistant Enterobacterales, detected by reduced susceptibility to any carbapenem (usually ertapenem), in isolates not known to have chromosomally-mediated mechanisms for reduced susceptibility, XDR: extensively drug resistant, classified using a standard definition [14]. *Including Candida spp. with established fluconazole resistance, C. krusei and C. glabrata, which were the only isolates in which fluconazole resistance was found. No C. auris isolates were detected in this setting during the period of study.