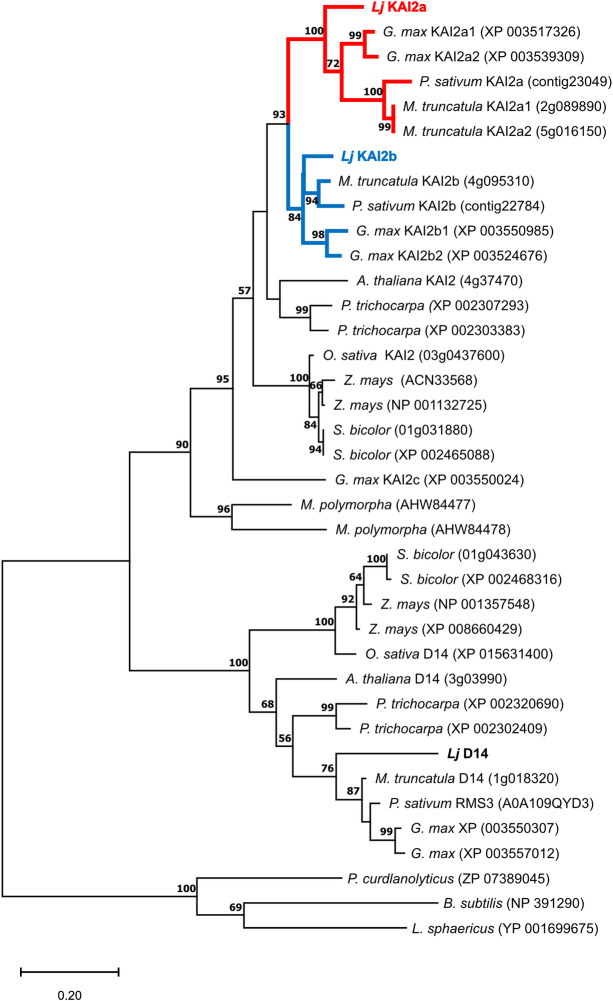
Fig 1. The KAI2 gene underwent duplication prior to diversification of the legumes.
Phylogenetic tree of KAI2 and D14 rooted with bacterial RbsQ from indicated species (Lotus japonicus; Glycine max; Pisum sativum; Medicago truncatula; Arabidopsis thaliana; Populus trichocarpa; Oryza sativa; Zea mays; Sorghum bicolor; Marchantia polymorpha). MEGAX was used to align the protein sequences with MUSCLE and generate a tree inferred by Maximum Likelihood method [72]. The tree with the highest log likelihood (-7359.19) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Values below 50 were ignored. KAI2 duplication in the legumes is highlighted by red and blue branches.