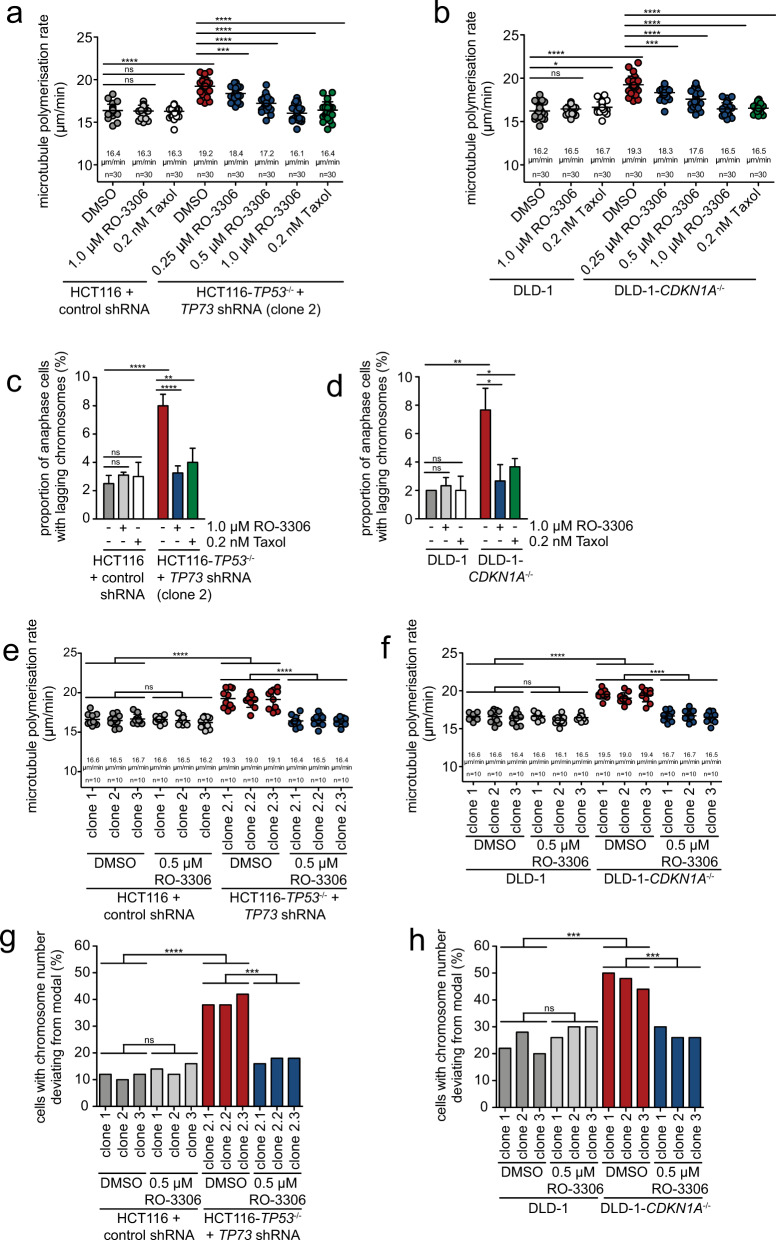
Fig. 4. Inhibition of CDK1 suppresses abnormal microtubule polymerization rates and chromosomal instability after loss of p53/p73 or p21CIP1.
a Rescue of abnormal microtubule growth rates upon partial CDK1 inhibition in TP53/TP73-deficient cells. HCT116 and TP53/TP73-deficient cells were treated with the CDK1 inhibitor RO-3306 or with low-dose Taxol for 16 h and mitotic microtubule plus end assembly rates were measured. Scatter dot plots show average microtubule polymerization rates (20 microtubules/cell, n = 30 mitotic cells from three independent experiments, mean ± SD, t-test). b Rescue of abnormal microtubule growth rates upon partial CDK1 inhibition in CDKN1A-deficient cells. DLD-1 and DLD-1-CDKN1A−/− cells were treated as in a and mitotic microtubule polymerization rates were determined (20 microtubules/cell, n = 30 mitotic cells from three independent experiments, mean ± SD, t-test). c Suppression of lagging chromosomes upon partial CDK1 inhibition in TP53/TP73-deficient cells. HCT116 and TP53/TP73-deficient cells were treated with the CDK1 inhibitor RO-3306 or with low-dose Taxol. The graph shows the proportion of cells exhibiting lagging chromosomes during anaphase (n = 300 anaphase cells from three independent experiments, mean ± SD, t-test). d Suppression of lagging chromosomes upon partial CDK1 inhibition in CDKN1A-deficient cells. Parental and CDKN1A-deficient DLD-1 cells were treated as in c. The graph shows the proportion of cells exhibiting lagging chromosomes during anaphase (n = 300 anaphase cells from three independent experiments, mean ± SD, t-test). e, f Mitotic microtubule growth rates in single-cell clones derived from control, TP53/TP73-deficient HCT116 cells or from parental and CDKN1A-deficient DLD-1 cells and treated with or without CDK1 inhibitor (RO-3306). Scatter dot plots show average microtubule growth rates (20 microtubules/cell, n = 10 mitotic cells for each clone, mean ± SD, t-test). g, h Chromosome number variability in single-cell clones derived from control, TP53/TP73-deficient HCT116 cells or from parental and CDKN1A-deficient DLD-1 cells and treated with or without CDK1 inhibitor. The proportion of cells with a chromosome number deviating from modal (45 chromosomes for HCT116, 46 chromosomes for DLD-1) was calculated for three independent clones (n = 50 cells per condition).