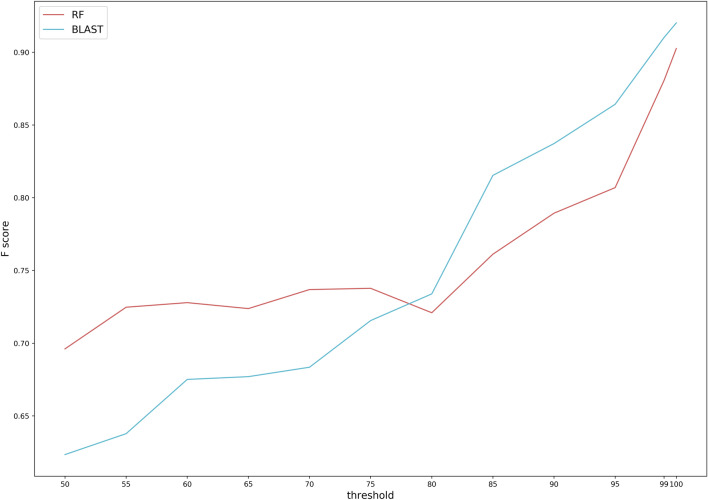
Figure 8.
Computed F1 scores of the final Random Forest (RF) model and BLASTp in a Leave-One-Group-Out Cross-Validation (LOGOCV) across different thresholds for sequence similarity. Our final predictive model was compared with BLASTp using a LOGOCV scheme. In every round, a group was held out that was controlled by sequence similarity. The model was trained on all other sequences, after which the bacterial hosts related to the held-out group were predicted. The sequences in this group were also subjected to a local BLASTp search via BioPython against the database without the held-out group36. The LOGOCV was repeated for different thresholds of sequence similarity in the dataset that controlled the grouping in the cross-validation (i.e. the lower the threshold, the more sequences were grouped into the held-out group, leaving fewer sequences to train the model with or perform a BLASTp search against). At every threshold, the F1 score was computed for predictions made by the RF model and by BLASTp.