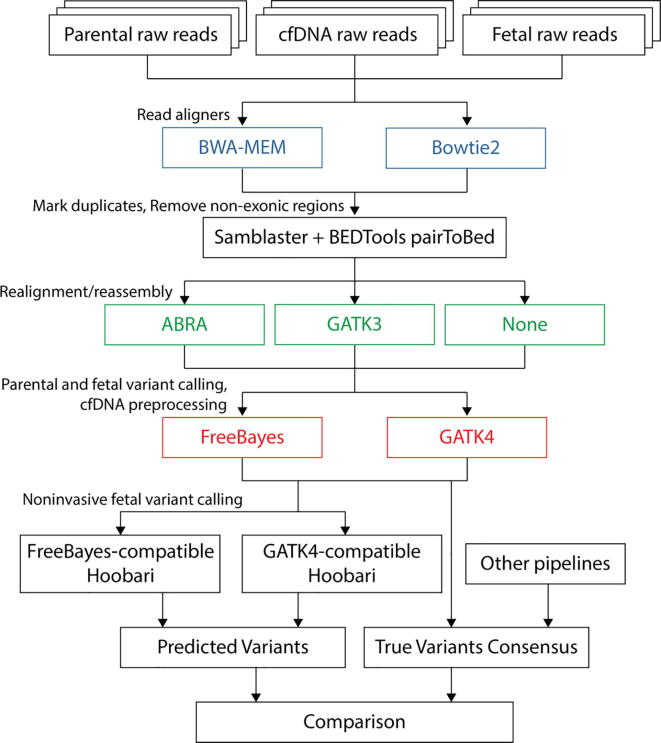
Fig. 1.
Experimental workflow for comparing fetal genotyping pipelines. This is the workflow for comparing pipelines for noninvasive fetal variant callers. Twelve pipelines were tested, based on possible combinations of two read aligners, three post-alignment approaches, and two variant calling programs. Each pipeline begins with raw sequence FASTQ files of the parents, the true fetal sample, and cfDNA. Alignment is performed using either BWA-MEM or Bowtie2 (Blue). Duplicate reads are removed using Samblaster. Non-exonic regions are sliced out using BEDTools pairToBed, by keeping only read pairs that have at least one read covering an exonic region. ABRA and GATK3 IndelRealign are compared against avoidance of any realignment or reassembly (Green). FreeBayes and GATK4 are used for variant calling of the parents and the true fetal samples, and for preprocessing of the cfDNA sample (Red). Eventually, FreeBayes- or GATK4-comparitble Hoobari is run to call fetal variants using the cfDNA reads and the parental genotypes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)