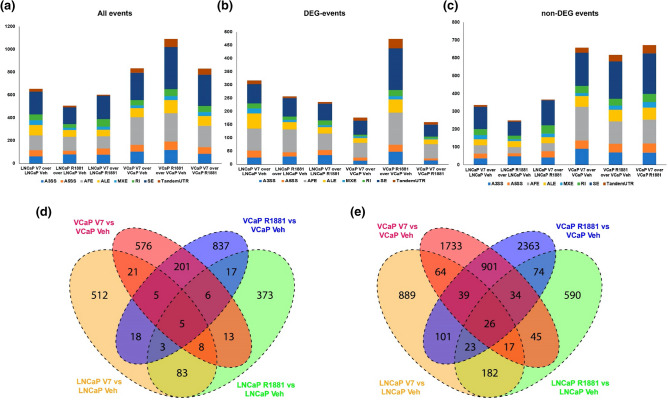
Figure 2.
Analysis of the alternative splicing events in the LNCaP and VCaP models. The number of each type of event derived from the reads of RNA-seq data as described in methods of LNCaP treated with R1881 (AR full length), VCaP treated with R1881, LNCaP- AR-V7 treated with Doxycycline and VCaP-AR-V7 treated with Doxycycline relative to the corresponding vehicle. (a) All events (total); (b) events in regulated genes (DEG), and (c) events in non-regulated genes (non-DEG). A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; AFE, alternative first exons; ALE alternative last exons; MXE, mutually exclusive exons; RI, retained introns; SE, skipped exons; Tandem UTR, Tandem 3′UTRs. (d) Overlap of splicing events in the two cell lines and for each AR isoform studied with Bayesian Factor (BF) ≥ 3 and changes in fraction isoform abundance (ψ) between treatment conditions greater than 0.2 (Δψ > 0.2). (e) Overlap of splicing events in the two cell lines and for each AR isoform studied with Bayesian Factor (BF) > 3 and no cut-off for change in fraction isoform abundance (ψ) between treatment conditions (Δψ = 0).