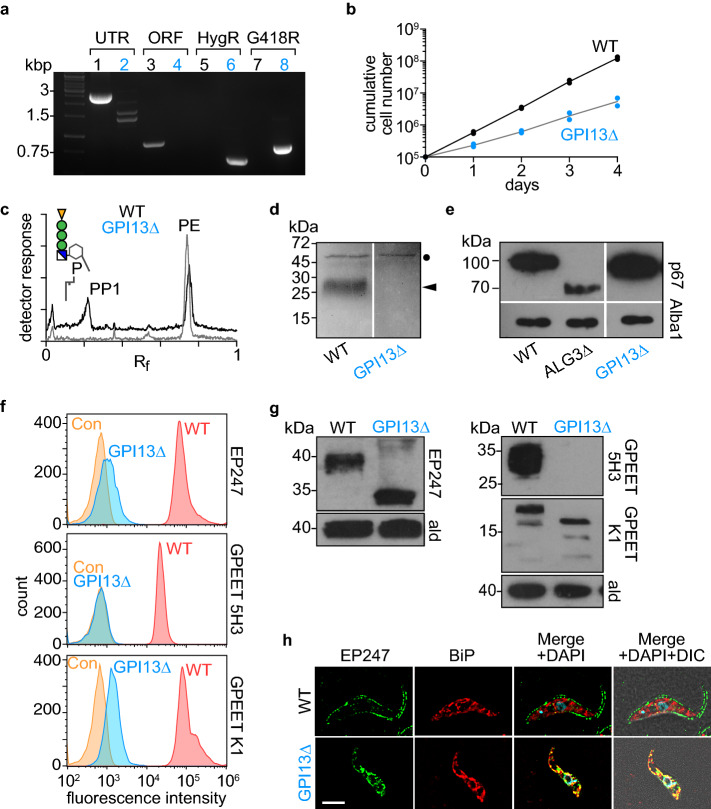
Figure 5.
Evaluating Gpi13 as the M5-DLO scramblase in procyclic form Trypanosoma brucei. ( a) PCR confirmation of TbGPI13 gene replacement using primers specific to the TbGPI13 UTR and ORF, and HygR/G418R gene replacement cassettes. Lanes 1, 3, 5 and 7 correspond to wild-type cells, whereas lanes 2, 4, 6 and 8 correspond to TbGPI13 knockout cells. (b) Growth of TbGPI13 knockout (GPI13Δ) and wild-type (WT) cells. Data points represent mean values from 2 independent experiments. (c) Thin layer chromatographic analysis of polar lipid extracts from [3H]-ethanolamine-labeled WT (top trace) and GPI13Δ (bottom trace) cells. The chromatograms were visualized using a radioactivity scanner. The WT trace is displaced upwards for clarity. PP1 and PE refer to the major GPI precursor in procyclic trypanosomes (structure shown in schematic) and PE, respectively. (d) SDS-PAGE/fluorography of protein extracts from [3H]-ethanolamine-labeled WT and GPI13Δ cells showing loss of GPI-anchored GPEET procyclin (indicated by arrow head) in GPI13Δ cells. The band at ~ 50 kDa (indicated by filled circle) represents eukaryotic elongation factor 1A, a protein that is
modified by ethanolamine phosphoglycerol and therefore metabolically labeled with [3H]-ethanolamine. (e) Analysis of p67 N-glycosylation by anti-p67 immunoblot of whole cell protein samples from WT and GPI13Δ cells. A sample from a known N-glycosylation defective cell line (ALG3Δ) was used as a positive control. Alba1 was probed using anti-Alba1 antibody and used as a loading control. (f) Flow cytometry analysis of surface expression of GPI-anchored EP and GPEET procyclins in WT and GPI13Δ cells. The middle and bottom panels correspond to flow cytometry analysis of GPEET using 5H3 antibody (middle) and K1 antiserum (bottom). Control (Con) samples were generated by omitting the primary antibody. (g) Anti-EP and anti-GPEET (K1) procyclin immunoblots of whole cell samples from WT and GPI13Δ cells. Aldolase was probed using anti-aldolase antibody and used as a loading control. (h) WT and GPI13Δ cells were fixed and stained with antibodies against EP and BiP (as a marker for the endoplasmic reticulum) in combination with fluorophore-conjugated secondary antibodies and analyzed by fluorescence microscopy. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) in the merged panels. DIC, differential interference contrast. Scale bar = 5 µm.