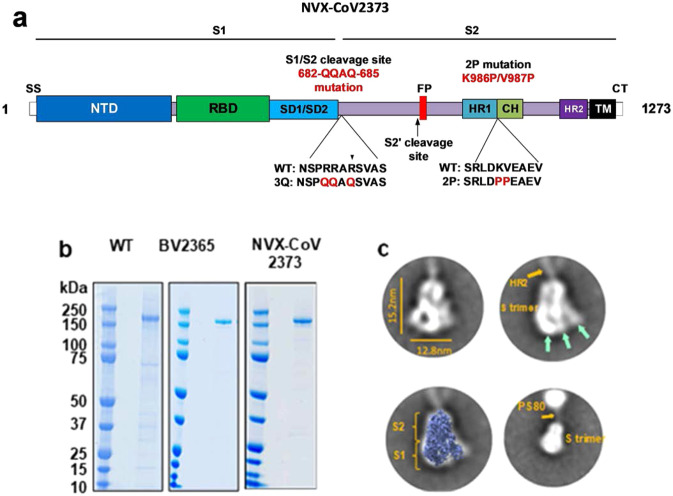
Fig. 1. SARS-CoV-2 spike glycoprotein constructs.
a Linear diagram of the full-length SARS-CoV-2 spike (S) protein showing the S1 and S2 subunits. Structural elements include a cleavable signal sequence (SS, white), N-terminal domain (NTD, dark blue), receptor binding domain (RBD, green), subdomains 1 and 2 (SD1/SD2, light blue), fusion peptide (FP, red), heptad repeat 1 (HR1, yellow), central helix (CH, light green), heptad repeat 2 (HR2, purple), transmembrane domain (TM, black), and cytoplasmic tail (CT, white). The native furin cleavage site was mutated (RRAR→QQAQ) to be protease resistant to generate the full-length BV2365 variant. BV2365 was further stabilized by introducing two proline (2P) substitutions at positions K986P and V987P to produce the double mutant NVX-CoV2373. b Reduced SDS-PAGE gel of purified full-length wild-type (WT), BV2365, and NVX-CoV2373 (representative of 3 to 10 lots). WT spike is produced as a mixture of cleaved and uncleaved proteins. Figure shows purified uncleaved WT spike protein. c Transmission electron microscopy and 2D class averaging of NVX-CoV2373. 2D images were constructed from 28,623 NVX-CoV2373 particles followed by two rounds of 2D averaging. 2D images of NVX-CoV2373 S-trimers showing well-defined triangle-shaped particles with a length of 15 nm and a width of 12.8 nm. The S1 apical surface with the N-terminal receptor and receptor-binding domain (NTD/RBD) is indicated by green arrows. Faint protrusions (orange arrow) extending from the tip of the trimers were evident and appear to correspond to the S2 HR2 domain. Class average images showing a good fit of NVX-CoV2373 S-trimer with cryoEM solved structure of the SARS-CoV-2 trimeric spike protein ectodomain (EMD ID: 21374) overlaid on the 2D image. 2D class averaging using a larger box size showing 2D class average image with the less well-defined HR2 (orange arrow) anchoring the S-trimer to polysorbate 80 (PS80) micelle by the C-terminal TM. Source data are provided as a Source Data file.