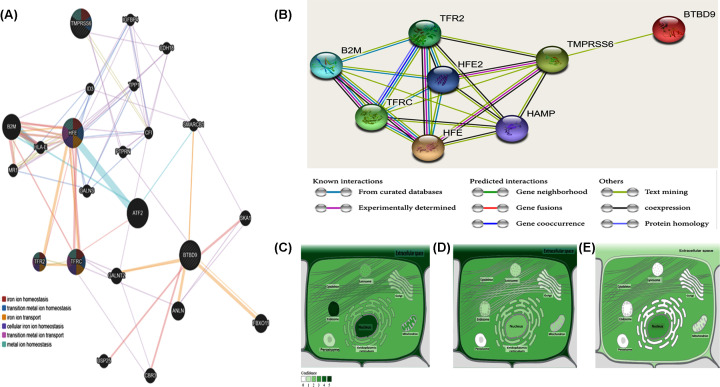
Figure 1. In silico data analysis.
(A) Gene–gene functional interaction network generated by GeneMANIA. The network nodes have been colored by function (i.e. gene ontology annotation) (data source: https://genemania.org/search/homo-sapiens/HFE/TMPRSS6/BTBD9). (B) Protein–protein interaction using STRING version 11. Each network node represents the protein produced by a single locus. Edges represent protein–protein associations (i.e., contribute to a shared function; this does not necessarily mean they are physically binding to each other). The line colors of the edges indicate the type of interaction evidence that is explained in the figure key. The Network showed a relationship with iron-metabolism related proteins (data source: https://string-db.org). (C–E) Subcellular localization of HFE, TMPRSS6, and BTBD9 proteins. Darker color, according to the provided color key, is indicating more abundance (data source: https://compartments.jensenlab.org). Abbreviations: ANLN, anillin actin-binding protein; ATF2, activating transcription factor 2; B2M, beta-2-microglobulin; BTBD9, BTB domain containing 9; CBR3, carbonyl reductase 3; CDH18, cadherin 18; CFI, complement factor I; FBXO11, F-box protein 11; GALNS, galactosamine (N-acetyl)-6-sulfatase; GALNT2, polypeptide N-acetylgalactosaminyltransferase 2; HAMP, Hepcidin; HFE, homeostatic iron regulator; ID3, inhibitor of DNA binding 3; IGFBP4, insulin-like growth factor binding protein 4; PTPRN, protein tyrosine phosphatase, receptor type N; SMARCB1, SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily b, member 1; SKA1, spindle and kinetochore associated complex subunit 1; TFR2, transferrin receptor 2; TFRC, transferrin receptor; TMPRSS6, transmembrane protease, serine 6; TPP1, tripeptidyl peptidase 1; USP25, ubiquitin specific peptidase 25; major histocompatibility complex, class I-related, HLA-E, major histocompatibility complex, class I, E; HLH protein.