Figure 1.
Features of Kinase Target Interaction and Pipeline for SDR Identification
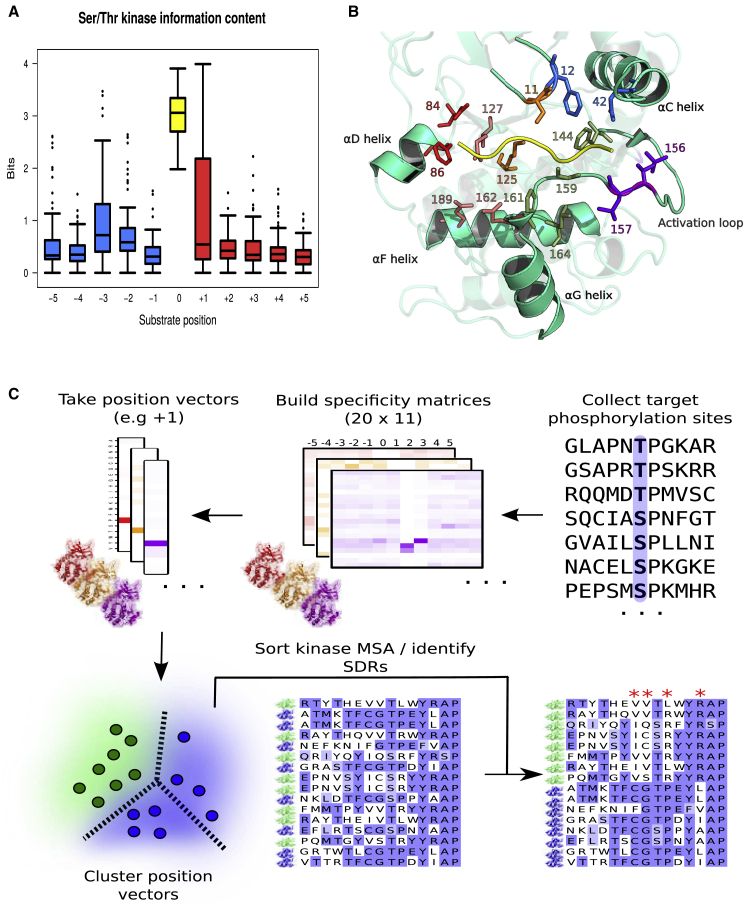
(A) Sequence constraint for substrate positions −5 to +5 for 119 serine/threonine kinases, measured as the bit value for the corresponding column of the kinase PWM.
(B) Interface between a protein kinase (human protein kinase A, PDB: 1ATP) and substrate peptide at the substrate-binding site (Zheng et al., 1993). Kinase residues that commonly bind the substrate peptide (yellow) are represented in stick format and colored according to the corresponding substrate position (−3: red, −2: pink, −1: orange, +1: green, +2: blue, +3: purple). Residue numbering represents the relevant positions of the Pfam protein kinase domain (PF00069).
(C) Semi-automated pipeline for the inference of putative kinase SDRs (specificity-determining residues). The first step involves the construction of many kinase PWMs from known target phosphorylation sites. Vectors corresponding to a substrate position of interest (e.g., +1) are then retrieved from each PWM. An unsupervised learning approach (i.e., clustering) identifies kinases with a common position-based preference (e.g., for proline at +1). Alignment positions that best discriminate kinases belonging to 1 cluster from all others are then identified using computational tools for SDR detection.