Figure 5.
Evolution of Specificity for Orthologous Kinases
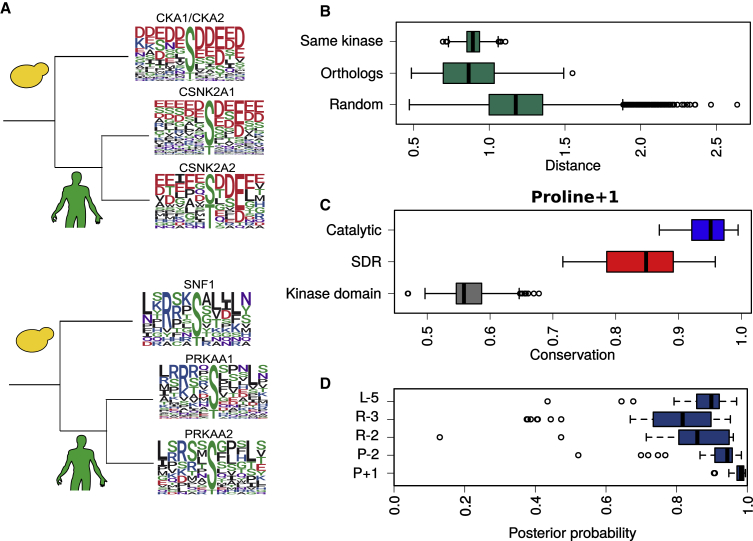
(A) Human and yeast kinase specificity logos for 2 different orthologous groups.
(B) Distribution of matrix distances between PWMs generated from phosphosite subsamples of the same kinase (top), orthologous yeast and human/mouse pairs (center), and random human-yeast pairs (bottom).
(C) Conservation of domain residues, SDRs, and catalytic residues for the proline+1 specificity. Each data point represents the average conservation (among kinase domain positions, SDR, or catalytic residues) for an alignment of orthologous kinases in which the human kinase is a predicted proline+1 kinase.
(D) Conservation of specificity for kinases orthologous to human kinases of predicted specificity (L-5, R-3, R-2, P-2, P+1). Each data point represents the average posterior probability (across all kinases in an orthologous group) that the specificity has been conserved.