Figure 3.
TMK1 Interacts with and Phosphorylates MAKR2 and Acts Upstream of MAKR2 in the Regulation of Root Gravitropism
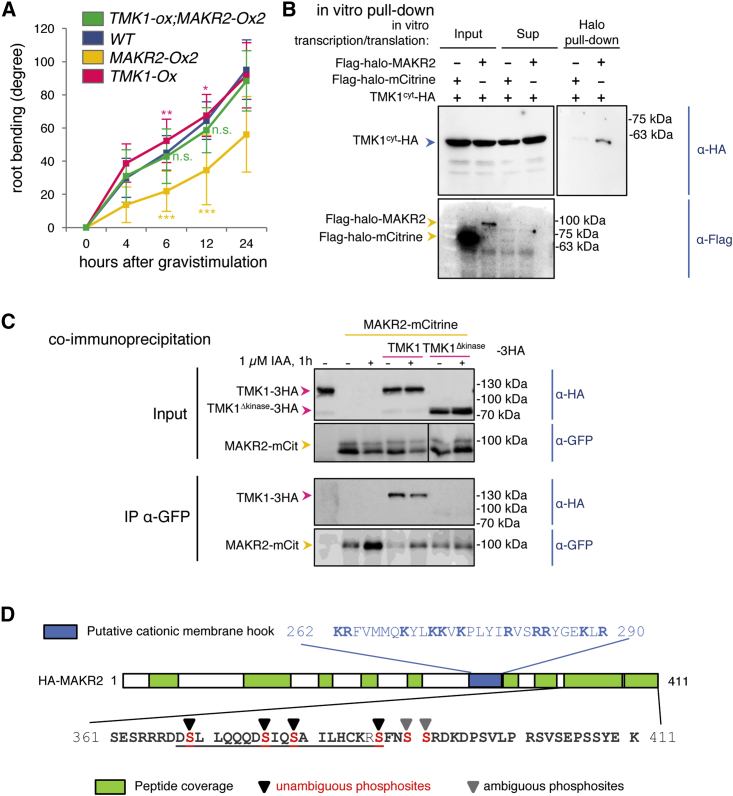
(A) Kinetics of root gravitropic bending after reorienting seedlings of the genotypes indicated in the top left corner at a 135° angle. See Figure S3C for a statistical comparison. A linear model was fitted on measurements from wild-type plants and the different mutants using lm() function from stats package available in R software (https://www.r-project.org/). This model estimates a weight for each variable (wild-type and mutant plants) and the associated probability that such weight is different from zero based on a t test. The probability derived from the t test is the p value in this comparison and significant differences were considered when p < 0.05.
(B) Pull-down assay using in-vitro-transcribed/translated proteins and Halo-tag purification. Co-purified proteins were visualized using an anti-HA antibody (labeled Halo pull-down). The inputs (labeled Inputs) and supernatant (labeled Sup) were tested to show the relative amounts of Halo- and HA-tagged proteins and the binding efficiency to HaloLink magnetic beads (as described in Yazaki et al.34). TMK1cyt corresponds to the isolated TMK1 cytoplasmic domain.
(C) Co-immunoprecipitation of full-length TMK1-3HA but not TMK1Δkinase-3HA with MAKR2-mCitrine. Immunoprecipitation (IP) of MAKR2-mCitrine with an anti-GFP antibody and immunoblotting (IB) using an anti-GFP antibody or anti-HA antibody. Protoplasts were incubated or not for 1 h with 1 μM IAA.
(D) The scheme represents the MAKR2 protein, with the peptides recovered by mass spectrometry highlighted in green and the phosphorylation sites shown by arrowheads (only found with active TMK1cyt but not inactive TMK1cyt-K616R). Black arrowheads indicate phosphorylation sites that could be determined with 100% accuracy, whereas gray arrowheads indicate ambiguity on which of the two consecutive serines is phosphorylated. The residues corresponding to the conserved C-terminal tail are underlined. The blue box indicates the position of the putative cationic membrane hook, and the corresponding Arg/Lys residues are highlighted in bold.
See also Figure S3.