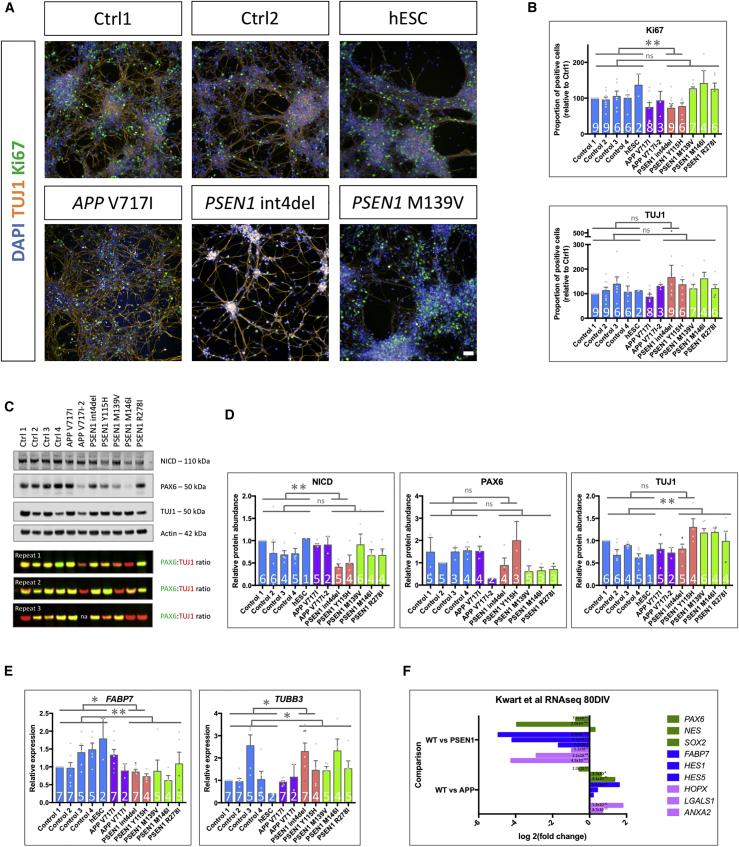
Figure 3.
Mutations in PSEN1 Reduce Notch Signaling and Lead to Premature Terminal Differentiation
(A) Representative images of day 27 iPSC-derived neural progenitors labeled for dividing cells (Ki67) and postmitotic neurons (TUJ1).
(B) High-content analysis and quantification of the relative proportion of proliferative (Ki67 positive) and neuronal (TUJ1 positive) cells.
(C and D) Representative western blot and quantification for the cleaved, active Notch intracellular domain (NICD), PAX6 neural progenitor marker, and TUJ1 neuronal marker. Note, due to variability in Ctrl1, PAX6 data were normalized to Ctrl2. Due to similar molecular weights, the ratio of PAX6 to TUJ1 intensity represents a measure of terminal differentiation. na, not available.
(E) qPCR analysis of the expression of the Notch readout gene FABP7 (alias BLBP) and TUBB3 (neuronal tubulin).
(F) RNA sequencing (RNA-seq) expression data from Kwart et al. (2019), demonstrating expression of neural stem cell (NSC) markers (PAX6, NES, and SOX2), Notch signaling readout genes (FABP7, HES1, and HES5), and adult NSC markers (HOPX, LGALS1, and ANXA2) (Berg et al., 2019; Edri et al., 2015) in 80 days in vitro (DIV) iPSC-derived neurons from wild type (WT) compared with isogenic PSEN1 or APP mutant lines. Adjusted p values are represented within the histogram.
Blue, control; purple, APP mutant cells; pink, mutations in the PSEN1 extracellular loop; green, mutations in PSEN1 transmembrane and intracellular domains. The number of independent neural inductions is shown within the histograms. Data from different iPSC clones are depicted by gray data points (APP V717I, PSEN1 int4del, and PSEN1 R278I). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 via ANOVA with post hoc Tukey’s analysis (normal distribution tested via the Shapiro-Wilk test), as indicated in Table S1. Scale bar represents 50 μm. Error bars represent standard error of the mean.