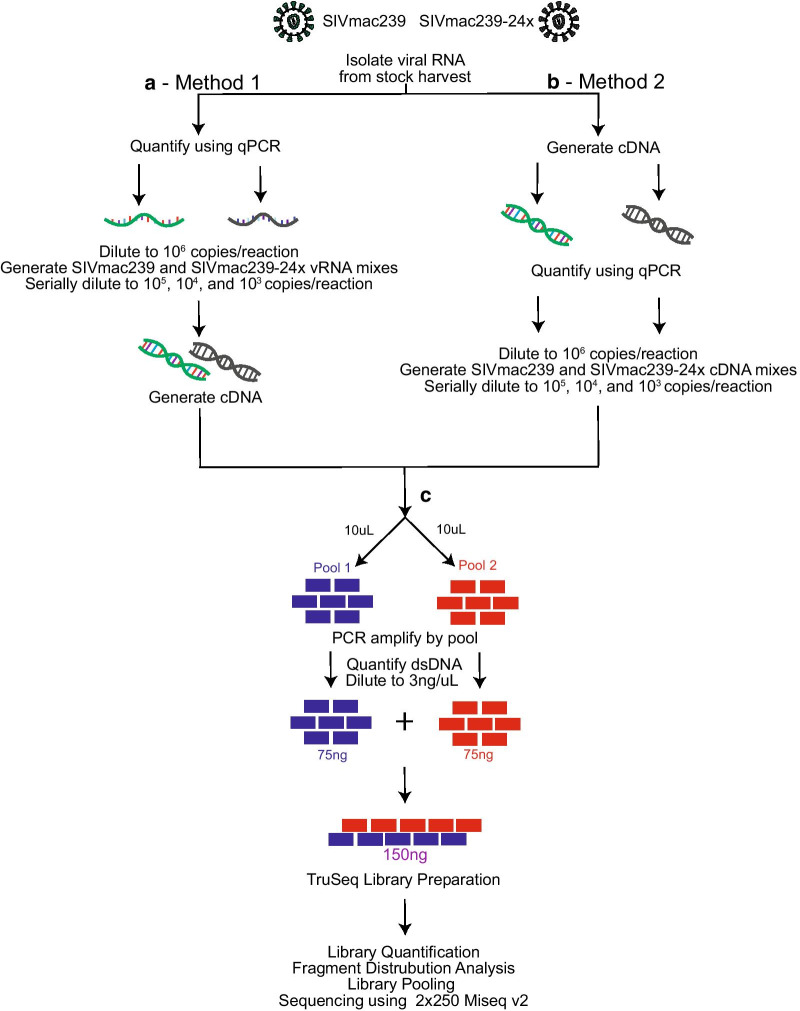
Fig. 3.
Schematic of experimental design. a Method 1: viral RNA was isolated from original stock and quantified via qRT-PCR. SIVmac239 and SIVmac239-24 × were diluted to 106 copies/reaction and mixed at the following SIVmac239:SIVmac239-24 × ratios: 100:0, 95:5, 90:10, 75:25, 40:60, and 0:100. Serial dilutions were preformed to 105, 104, and 103 copies per reaction. Viral cDNA was generated from viral RNA mixes, with one cDNA reaction per vRNA mix. b Method 2: viral RNA was isolated and approximately 107 viral RNA copies were added to each cDNA synthesis reaction. Viral cDNA copies were quantified using qRT-PCR and each was diluted to 106 copies per reaction. SIVmac239:SIVmac239-24 × mixes were generated at the following ratios: 100:0, 95:5, 90:10, 75:25, 50:50, and 0:100. cDNA mixes were then serially diluted to 105, 104, and 103 copies per reaction. c cDNA was used for multiplex PCR. PCR products were then combined at equimolar ratios and library prepped according to TruSeq Library Preparation documentation (Illumina). Libraries were quantified, pooled, and sequenced using a 2 × 250 v2 MiSeq cartridge