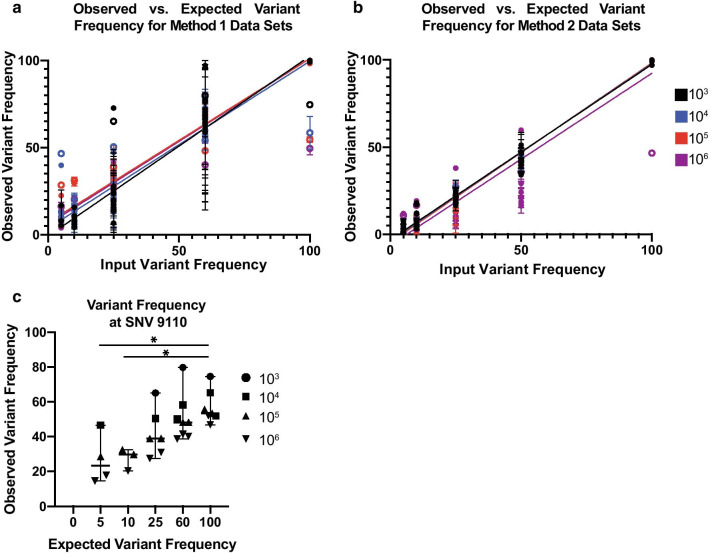
Fig. 4.
a Observed versus expected variant frequencies identified in the Method 1 (vRNA) mixed data sets. Observed variant frequency indicates percent SIVmac239-24× identified. Error bars indicate standard deviation for each replicate. Linear regressions colored by vRNA templates. Open circles indicate SNV 9110. b Observed versus expected variant frequencies identified in Method 2 (cDNA) mixed data sets. Observed variant frequency indicates percent SIVmac239-24× identified. Error bars indicate standard deviation for each replicate. Linear regressions colored by cDNA templates. Open circles indicate SNV 9110. No significant difference is observed between input templates and slope. c Observed versus expected variant frequency for SNV at 9110 for vRNA data sets. Lines represent medians and error bars indicate 95% confidence interval. Asterisk indicate p-value less than 0.05 as determined by Kruskal–Wallis