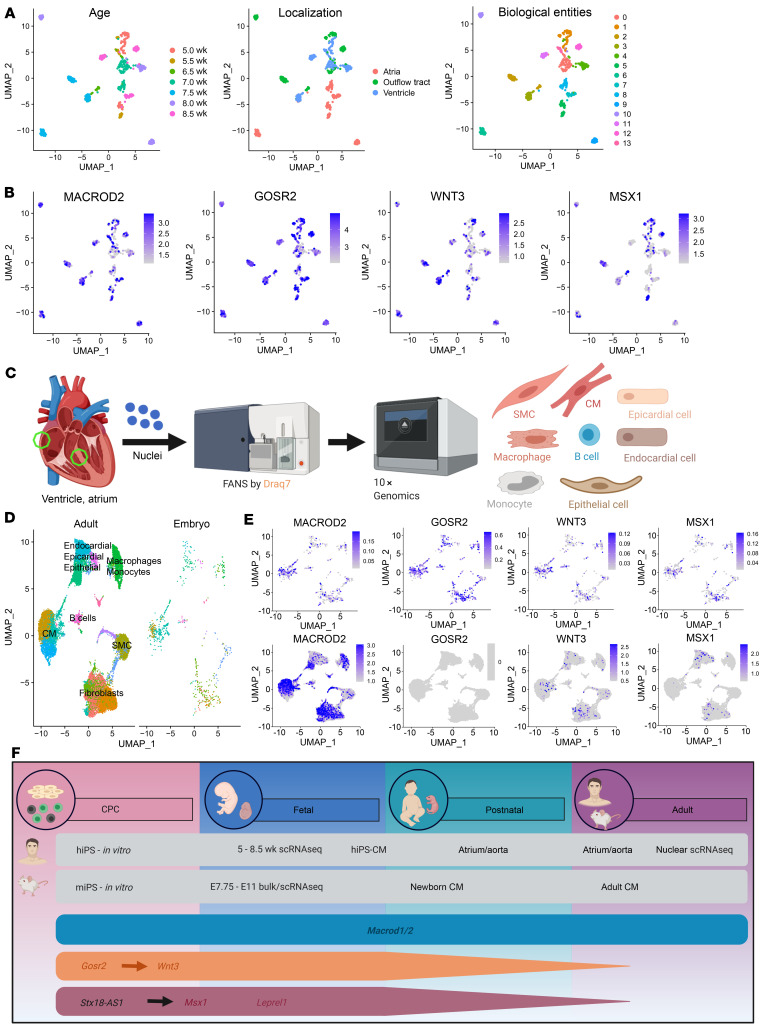
Figure 7. Role of SNP-carrying candidate genes in human cardiac development.
(A) Unbiased clustering of embryonic cells (n = 669) into biological entities. Cells are labeled on the basis of age as well as anatomical localization for purposes of visualization. (B) Relative expression of MACROD2, GOSR2, WNT3, and MSX1 in embryonic heart cells. (C) Schedule of scRNA-Seq analysis of cells from atria and ventricles (n = 17,782). (D) Clustering of embryonic and adult cells and identification of cell types. (E) Expression of candidate genes in the integrated data set split by embryonic cells (upper panel) and adult cells (lower panel). (F) Expression of candidate genes associated with TGA (turquoise), ATAV (orange), or septal defects (red) in vitro and in vivo during different stages of the developing murine and human heart. hiPS, human iPSCs; miPS, murine iPSCs; SMC, smooth muscle cells.