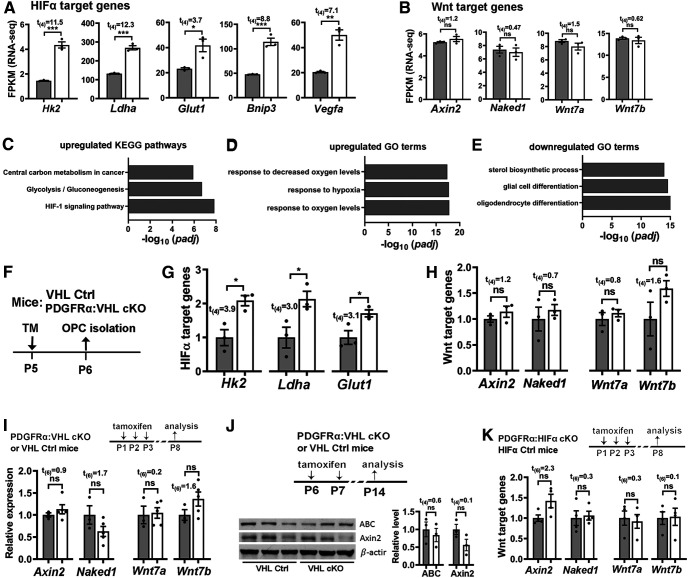
Figure 6.
Stabilizing HIFα activates HIFα-mediated signaling but does not perturb Wnt/β-catenin signaling in the oligodendroglial lineage and in the CNS. A, B, Relative expression of HIFα and Wnt target genes from unbiased RNAseq analysis. FPKM, Fragments per kilobase of transcript per million mapped reads, a normalized measurement of gene expression. The RNA was prepared from the spinal cord of P5 Sox10-Cre:Vhlfl/fl mice (n = 3, white bars) and littermate control (Vhlfl/+ or Vhlfl/fl; n = 3, gray bars) mice. C–E, Top three significant enriched KEGG Pathway and GO terms among 255 upregulated and 351 downregulated genes derived from unbiased RNAseq. Note that HIFα signaling pathway (C) and response to hypoxia (D) were significantly upregulated, validating HIFα hyperactivation in Sox10-Cre:Vhlfl/fl mice and that oligodendroglial cell differentiation was significantly downregulated (E), which is in agreement with the data of Figure 2A–D. F, Experimental design for G and H. PDGFRα:VHL cKO (n = 3, white bars) and littermate control (Vhlfl/fl, n = 3, gray bars) mice were treated with TM at P5, and forebrain OPCs at P6 were acutely purified by MACS. G, H, qRT-PCR quantification of HIFα target gene (G) and Wnt target gene (H) expression in purified OPCs. I, Expression of Wnt target gene (Axin2 and Naked1) and Wnt7a/7b in P8 spinal cord (n = 3, Ctrl, gray bars; n = 5, PDGFRα cKO, white bars) quantified by qRT-PCR. J, Western blot images and relative expression (normalized to the internal loading control β-actin) of the active form of β-catenin (ABC) and Axin2 in P14 forebrain (n = 3 each group). K, Expression of Wnt target genes (Axin2 and Naked1) and Wnt7a/7b in P8 spinal cord measured by qRT-PCR (n = 4 each group).