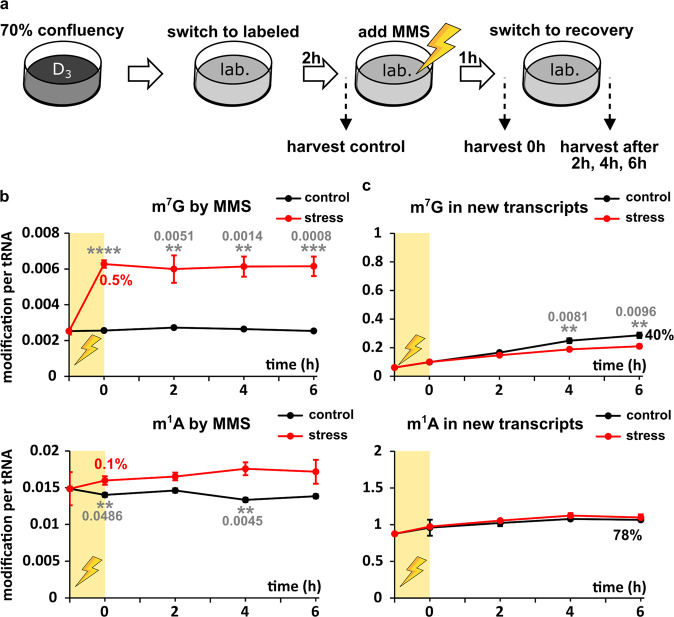
Fig. 6. Effect of methylation stress on tRNA modifications.
a 70% confluent CD3-methionine labeled cells were incubated with fully labeled media for 2 h before the LD50 dose of methyl methanesulfonate (MMS, yellow shaded area) was added. Control samples were treated the same, substituting the MMS stock solution with PBS. After 1 h the stress (or control) media was replaced by fresh labeled media. After set time points, cells were harvested and tRNAPheGAA was purified and subjected to LC-MS/MS analysis. b Unlabeled modifications were referenced to unlabeled canonicals to calculate the amount of modifications arising from direct methylation damage by MMS. Red numbers at time point 0 give the percentage of damaged nucleoside referenced to the naturally occurring amount of the respective modification (amount of original modification before experiment initiation). c Labeled modifications were referenced to labeled canonicals to calculate the amount of modification in new tRNA transcripts. The numbers at time point 6 give the percentage of modification amount in the control sample referenced to the naturally occurring amount of the respective modification. All experiments are from n = 3 biol. replicates. Symbols reflect the mean and error bars reflect standard deviation. P-values from student t-test (equal distribution, two-sided): *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Where reasonable, exact P-values are given as grey numbers in the figure.