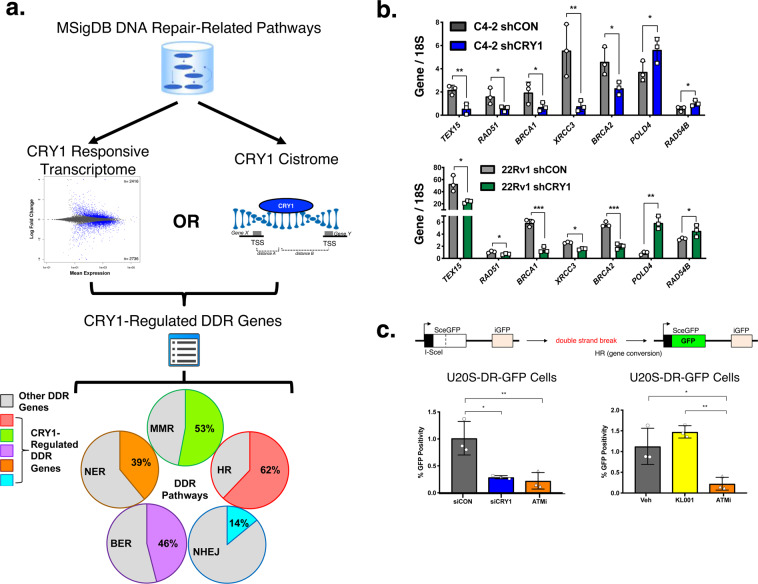
Fig. 5. CRY1 transcriptome and cistrome analyses identify direct regulation of DNA repair by CRY1.
a Schematic describing the comparison of RNA-Seq and ChIP-Seq datasets. Briefly, the DNA damage response transcripts regulated by CRY1 with p < 0.05 (no fold change cutoff) and DDR genes with a CRY1 binding site within a TSS of binding were identified and organized into specific DDR pathways. b CRY1 expression was knocked down in C4-2-shCRY1 and 22Rv1-shCRY1 cells for 72 h, RNA was harvested, and qPCR was performed. c CRY1 was knocked down in U20S-DR-GFP cells for 72 h via siRNA and cells were treated with ATM inhibitor for 24 h or with 10 µM KL001 for 48 h. Cells were harvested for flow cytometry. N = 3 independent experiments. Data are presented as mean values ± SEM and analyzed using two-way Anova (*p < 0.05, **p < 0.01, and ***p < 0.001). Statistical significance was evaluated at 0.05 alpha level with GraphPadPrism, version 8.3.1, Mac. Source data are provided in the Source Data file.