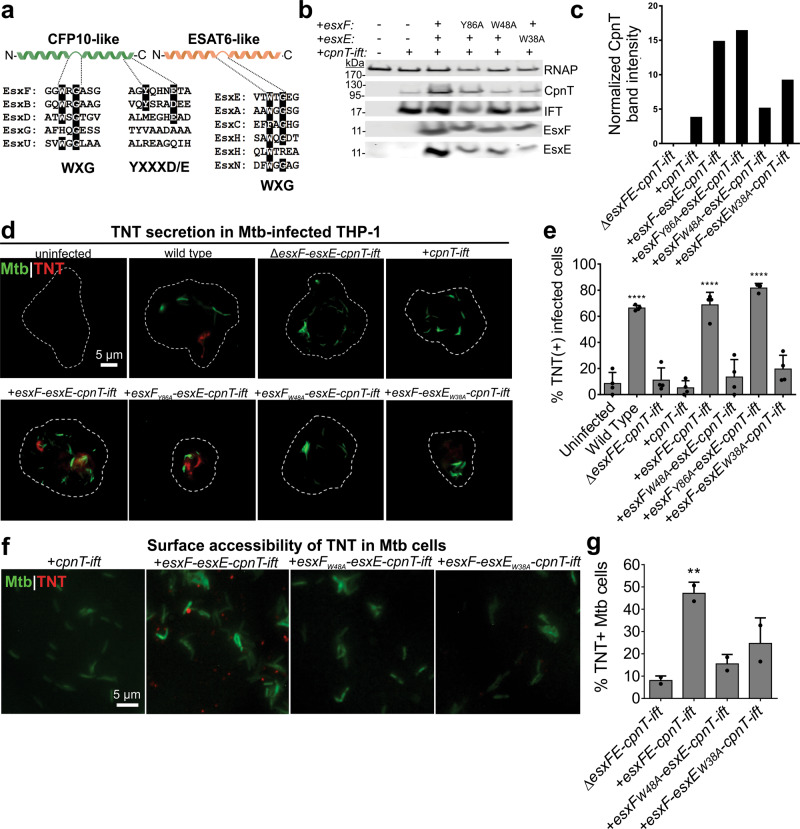
Fig. 2. The WXG motif is required for EsxE-EsxF function.
a Sequence alignment of the WXG YXXXD/E motifs in Esx heterodimer pairs of M. tuberculosis. b Immunoblot of Mtb whole-cell lysates probed with antibodies for the indicated proteins. RNA polymerase (RNAP) was used as a loading control. Representative of two experiments c Quantification of the CpnT bands from the immunoblot in b using ImageJ. d Detection of TNT in the cytosol of Mtb-infected macrophages. Representative fluorescence microscopy images of THP-1 macrophages 48 h after infection with DMN-trehalose-labeled Mtb. THP-1 cells were treated with digitonin to only permeabilize the plasma membrane and were probed with anti-TNT antibody (Alexafluor-594, red). Representative of four independent experiments. e Quantification of d. Infected macrophages were scored as TNT+ or TNT− based on the presence or absence of bright red punctae and quantified as % TNT-positive cells. At least 100 cells were analyzed of at least four independent experiments. Data shown is the mean ± SD from four independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnet’s multiple comparison test using the uninfected sample as the negative control. P < 0.0001. f Surface acccessibility of TNT of the indicated Mtb strains by fluorescence microscopy with DMN-trehalose (green), and probed with α-TNT and an Alexafluor-594-conjugated secondary antibody. Data are from at least ten fields of view from two independent experiments. g Quantification of f. Percentage of TNT-positive cells out of total bacteria from at least five fields of view. N ≥ 1000 bacterial cells total over two independent experiments. Shown is mean and standard deviation from two experiments. The statistical analysis was performed using one-way ANOVA analysis with Dunnet’s multiple comparison test using the ΔesxFE-cpnT-ift strain as the negative control. P = 0.016. Source data are provided in the Source data file.