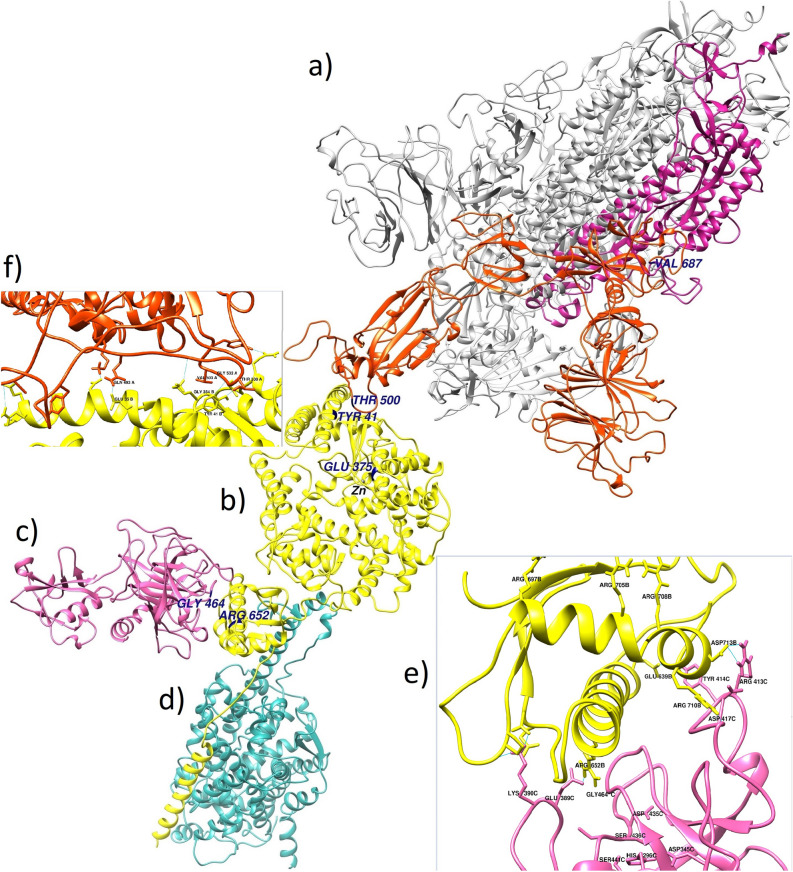
Figure 3.
Structural alignment of the Spike trimer, Spike monomer-ACE2, ACE2-TMPS2, and ACE2-B0AT1 complexes. The whole picture of the spatial positions of the main partners of ACE2 in the pathogenic process of SARS-CoV-2. This figure has been produced by the UCSF Chimera 1.14-linux_x86_64 (https://www.cgl.ucsf.edu/chimera/download.html). (a) The 3D structure of the Spike protein trimer, the S1 and S2 subunits presented in orange-red and violet-red colors, respectively. (b) The 3D structure of the ACE2 human receptor (yellow). (c) The 3D structure of the TMPRSS2 protease (hot pink); (d) The 3D structure of the B(0)AT1 (SLC6A19) Amino acid transporter (light sea green); (e) The interface of ACE2 and TMPRSS2 proteins; Letters B and C indicate ACE2 and TMPRSS2 chain IDs, respectively. Arg652 of ACE2 has been placed in the best position relative to the triad of TMPRSS2 (His296, Asp345, and Ser441) and its key residues (Asp435, Ser436, and Gly464). (f) The interface of ACE2 and Spike proteins; the main residues of ACE2 and Spike involved in the interaction were shown.