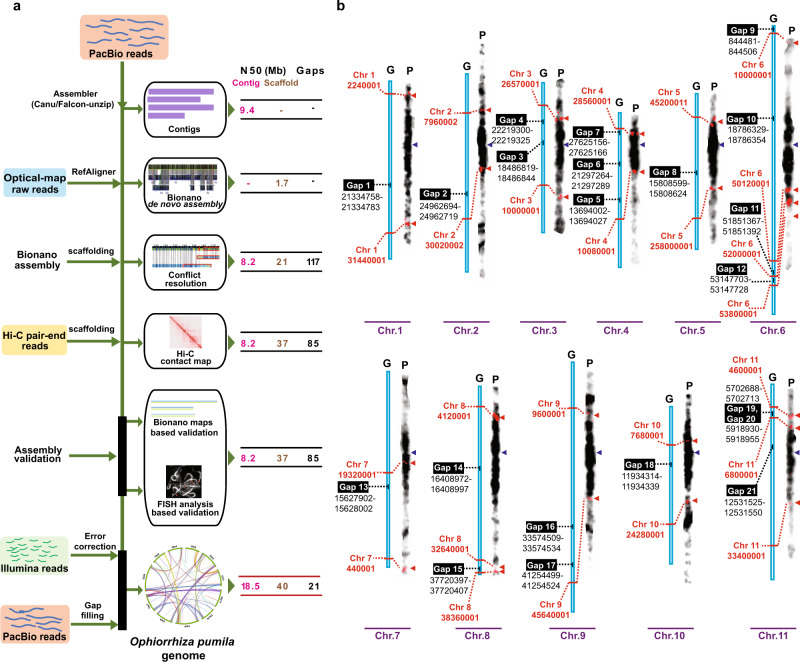
Fig. 1. Multistage assembly validation to achieve chromosome-level genome assembly for a high repeat content plant genome.
a The hierarchical genome assembly and assembly validation pipeline, and the improvement of assembly contiguity in terms of contig N50, scaffold N50, and assembly gaps of O. pumila genome. The assembly schema consistently corrects genome assembly from the point of contig-level assembly to the scaffold-level assembly, using orthogonal evidence and experimental validations. b Fluorescence in situ hybridization (FISH)-based validation of chromosome arms and orientation of scaffolds at the assembly gaps of Ophiorrhiza genome. The red triangle represents the site of FISH probe signal identified for a given chromosome, and the dotted line corresponds to the position at the scaffold. The purple colored triangle represents putative centromere at the pachytene chromosome, representing densely packed genomic segment, and hence darkly stained. The FISH experiment was repeated twice, and for each experiment, at least ten slides for each chromosome were observed and verified for signals as shown in b. G assembled chromosome, P pachytene chromosome with a FISH signal.