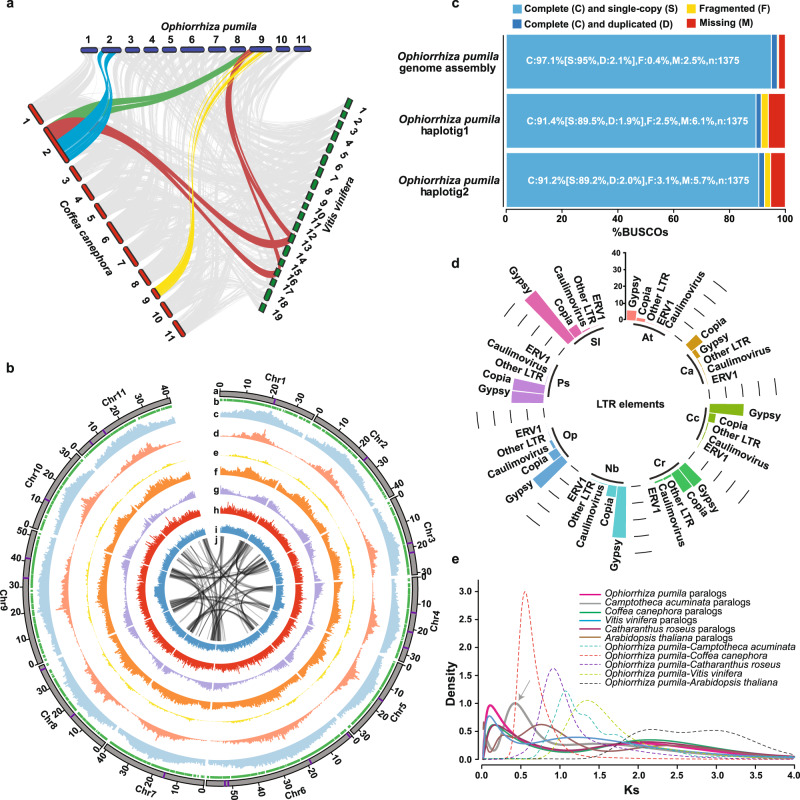
Fig. 3. O. pumila genomic landscape and convergent evolution of monoterpene indole alkaloid biosynthesis.
a Synteny blocks between O. pumila, C. canephora, and V. vinifera. Synteny analysis suggested karyotypic rearrangement between chromosomes 2 and 9 of O. pumila and C. canephora genomes, respectively. b Characteristics of the 11 chromosomes of O. pumila. Track a–c corresponds to chromosomes (assembly gap is depicted by purple line at each chromosome), phasing blocks, and repetitive sequences, respectively. Track d–i corresponds to the distribution of long terminal repeat (LTR)-Gypsy, distribution of LTR-Copia, GC density, distribution of predicted gene models, SNP density, and indel density, respectively. Track j corresponds to syntenic blocks. The bar, representing chromosomes, are scaled to chromosome lengths. c Evaluation of O. pumila genome assemblies using Benchmarking Universal Single-copy Orthologs (BUSCO) analysis. d Percentage of genome content comprising LTR elements for eight plant species. At Arabidopsis thaliana, Ca Camptotheca acuminata, Cc Coffea canephora, Cr Catharanthus roseus, Nb Nicotiana benthamiana, Op O. pumila, Ps Papaver somniferum, Sl Solanum lycopersicum. e Synonymous substitution rate (Ks) distribution plot for paralogs and orthologs of O. pumila with other eudicots as shown through colored continuous and dotted lines, respectively. The arrow highlights the recent whole-genome duplication identified in C. acuminata genome. Source data are provided as a Source data file.