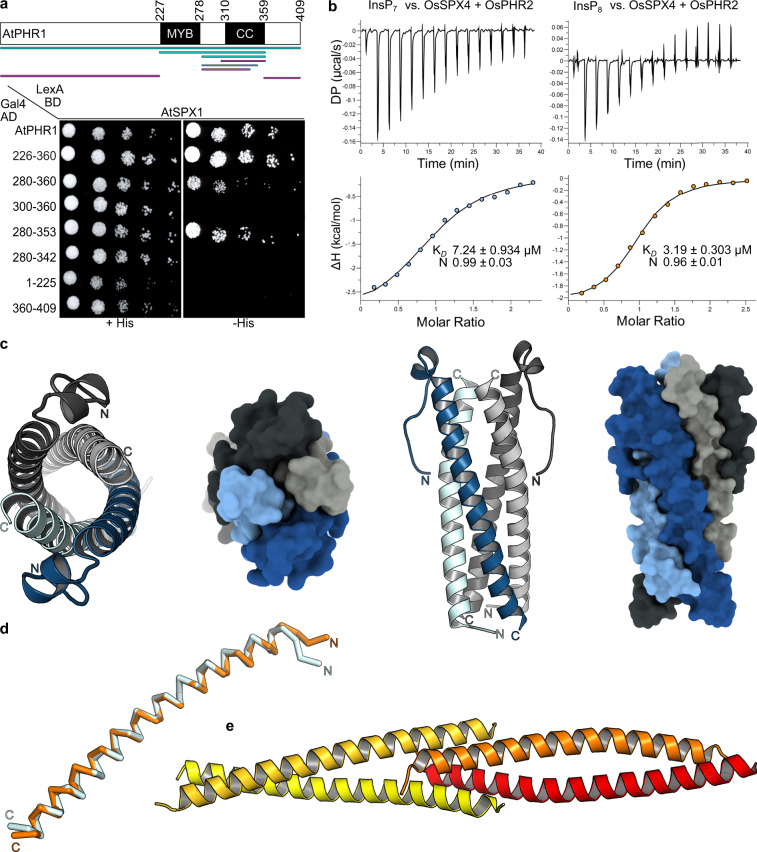
Fig. 1. AtSPX1 recognizes the AtPHR1 coiled-coil domain that crystallizes as a tetramer.
a Yeast co-expressing different AtPHR1 deletion constructs fused to the Gal4-activation domain (AD; prey) and full-length wild-type AtSPX1 fused to the LexA-binding domain (BD; bait) were grown on selective SD medium supplemented with histidine (+His; co-transformation control) or lacking histidine (−His; interaction assay) to map a minimal fragment of AtPHR1 sufficient for interaction with AtSPX1. Shown are serial dilutions from left to right. A schematic overview of the tested interacting (in cyan) and non-interacting (in magenta) AtPHR1 fragments is shown alongside (MYB, MYB–DNA-binding domain; CC, coiled-coil domain). b Isothermal titration calorimetry assays of InsP7 (400 µM 5PP-InsP5; left panel) and InsP8 (500 µM 1,5(PP)2-InsP4; right panel) binding to OsSPX4–OsPHR2 (30 µM), respectively. Raw heats per injection are shown in the top panel and the bottom panel represents the integrated heats of each injection, fitted to a one-site binding model (solid line). The insets show the dissociation constant (KD) and binding stoichiometry (N) (±fitting error). c Ribbon and surface diagrams of the AtPHR1 CC four-stranded anti-parallel tetramer. Helices contributing to the dimer interface are shown in light and dark blue, respectively. Corresponding, symmetry-related helices completing the tetramer are shown in light and dark gray. d Structural superposition of two AtPHR1 core CC helices (Cɑ trace, in light blue) and ScCtp1 (PDB-ID 4 × 01, in orange)44. R.m.s.d. is ~1 Å comparing 45 corresponding Cɑ atoms. e Ribbon diagram of the ScCtp1 dimer-of-dimers CC domain, with contributing helices colored from yellow to red.