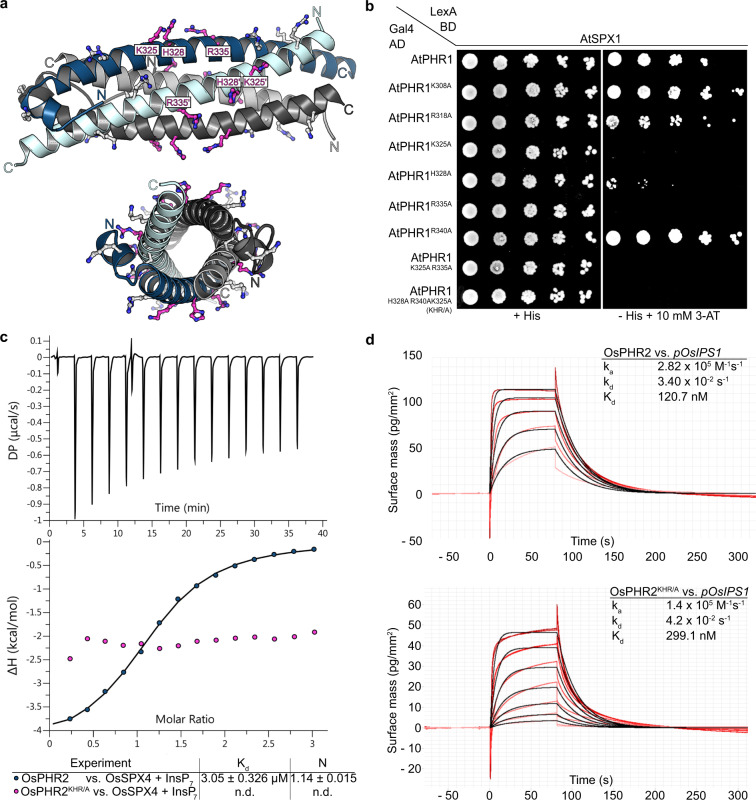
Fig. 3. The KHR motif at the surface of the PHR coiled-coil domain is required for the interaction with SPX domains.
a Ribbon diagram of the AtPHR1 CC domain with conserved basic residues located at the surface of the domain shown in bonds representation. The KHR motif (AtPHR1K325, AtPHR1H328, AtPHR1R335) is highlighted in magenta. b Mutational analysis of the basic residues in AtPHR1 CC. Yeast co-expressing AtPHR1226–360 variants in which surface-exposed basic residues have been replaced with alanine fused to the Gal4-AD (prey) and AtSPX1 fused to the LexA-BD (bait) were grown on selective SD medium supplemented with histidine (+His; co-transformation control) or lacking histidine and supplemented with 10 mM 3-amino-1,2,4-triazole (3-AT) (−His + 10 mM 3-AT; interaction assay) to identify residues required for interaction with AtSPX1 in yeast two-hybrid assays. Shown are serial dilutions from lift to right. c Isothermal titration calorimetry (ITC) assay of wild-type OsPHR2 and OsPHR2KHR/A (300 µM) vs. OsSPX4 (20 µM)–5PP-InsP5 (100 µM). Raw heats per injection are shown in the top panel, the bottom panel represents the integrated heats of each injection, fitted to a one-site binding model (solid line). The insets show the dissociation constant (KD) and binding stoichiometry (N) (± fitting error, n.d. no detectable binding). d Quantitative comparison of the interaction of OsPHR2 (top panel) or OsPHR2KHR/A (bottom panel) with pOsIPS1 by GCI. Sensorgrams show raw data (red lines) and their respective fits (black lines). The insets show summarize association rates (ka), dissociation rates (kd), and the dissociation constant (Kd) of the respective sample.