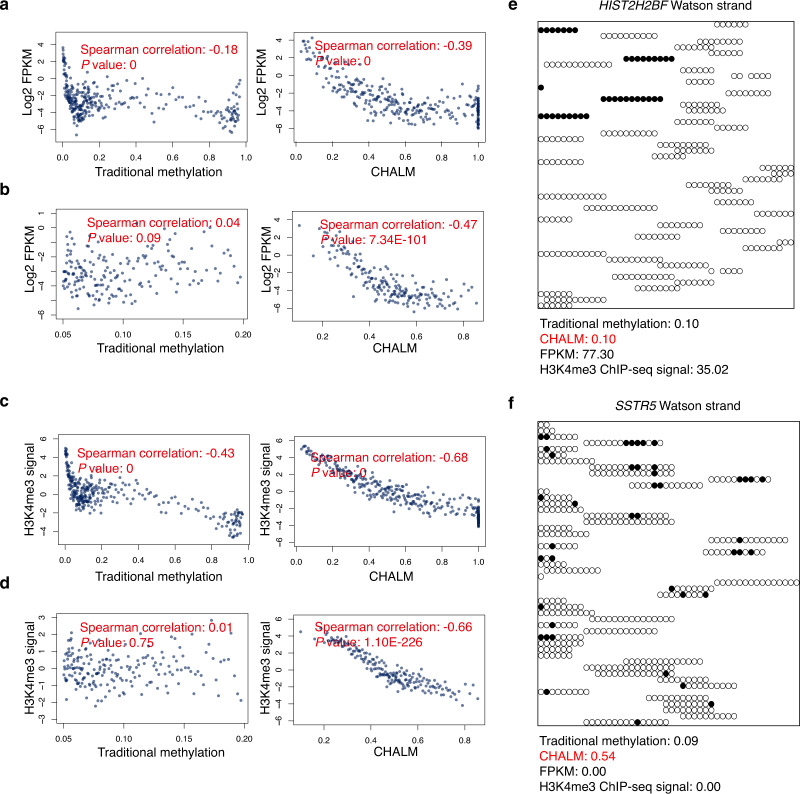
Fig. 2. The CHALM method better predicts gene expression.
a Scatter plots show the correlation between gene expression and methylation level calculated using both methods. Balanced promoter CGIs (Methods section) of CD3 primary cells are used. Each data point represents the average value of 10 promoter CGIs, and the Spearman correlation is calculated based on original data for each promoter CGI. Comparison of correlation (between the traditional method and CHALM) P values calculated by permutation (Methods section): <1 × 10−4. b Similar to a but focusing on low-methylation genes. Comparison of correlation permutation P values: <1 × 10−4. c Scatter plots show the correlation between H3K4me3 ChIP-seq intensity and methylation level calculated by the traditional and CHALM methods. Balanced promoter CGIs are used. Comparison of correlation permutation P values: <1 × 10−4. d Similar to c but focusing on low-methylation genes. Comparison of correlation permutation P values: <1 × 10−4. e, f Methylation status of reads mapped to the promoter CGI of HIST2H2BF or SSTR5, respectively. Black circles: mCpG; white circles: CpG.