figs2.
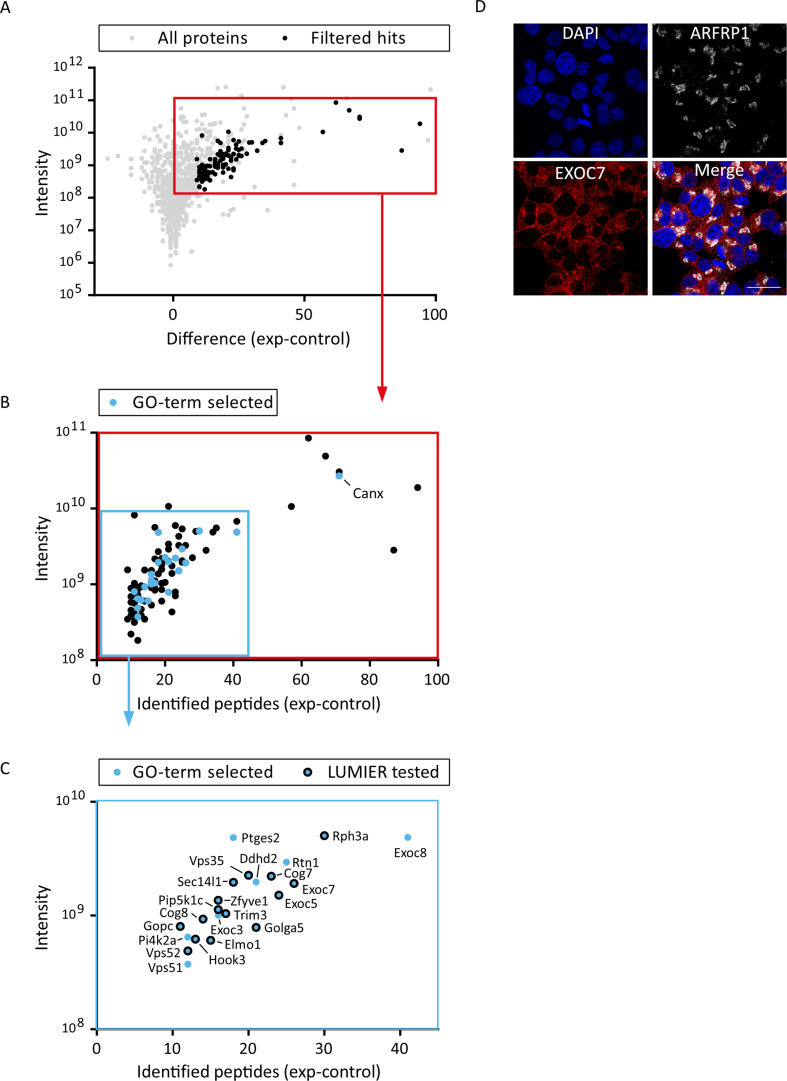
(A) Mass spectrometry analysis for GST-ARFRP1 pulldown. GTPγS-loaded GST-ARFRP1 and a GST control were used in pulldown experiments with mouse brain lysate and analyzed by mass spectrometry, using a spectral counting approach. Shown on the x-axis is the difference of the number of identified peptides for GST-ARFRP1 (exp) minus GST (control) for each protein group. On the y-axis is the total intensity of all peptides identified in exp and control. One thousand one hundred fifty-six protein groups were filtered for those containing at least 10 unique peptide IDs with a total intensity of ≥108 and having a peptide ID ratio (number of peptide IDs identified for GST-ARFRP1 over the total number of identified peptide IDs) of ≥0.9 (black dots). (B) Magnified view of the red box in A, showing the resulting 98 protein groups, which were further refined to proteins with a known Golgi-localization (GO:0005794) and/or an involvement in vesicular trafficking (GO:0016192) (blue dots). Calnexin (Canx) was not further pursued due to its known function in the endoplasmic reticulum. (C) Magnified view of the blue box in B. From the 23 GO-term selected protein groups, 16 were chosen as prey proteins for subsequent analysis in a LUMIER assay (blue dots with black outline). (D) Immunofluorescent pictures of Ins1 cells stained with indicated antibodies. Nuclei were visualized by DAPI. Scale bar represents 20 μm.