Figure 2.
Orthogonal identification of geldanamycin as a KDM4B inhibitor by high-content immunofluorescence imaging screen and a MALDI-FTICR mass spectrometry-based approach
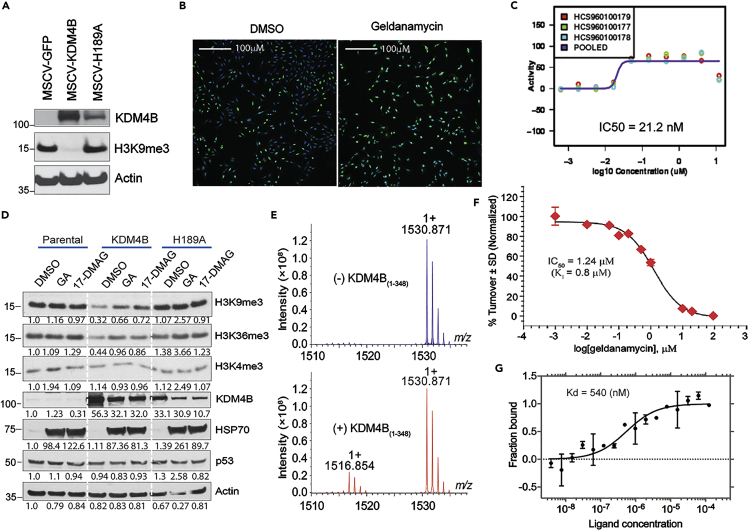
(A) Western blot for KDM4B expression in U2OS cells using an anti-KDM4B antibody.
(B) High-content immunofluorescence assay shows geldanamycin inhibits KDM4B activity. The U2OS cells expressing KDM4B were treated with 149nM of geldanamycin for 24 hr. The cells were fixed with 4% formaldehyde and permeabilized with 0.1% Triton X-100, followed by primary anti-H3K9me3 antibody incubation overnight at 4°C. The secondary goat α-rabbit-Alexa-488 and Hoechst 34,580 were added for imaging. The images were captured of each well at 10X using a GE Healthcare InCell 6,000 at 405nm to detect nuclear staining and 488 nm to detect H3K9me3. Green = H3K9me3, Blue = Hoechst 34,580.
(C) Dose-response curve shows IC50 of KDM4B inhibition by immunofluorescence assay. Purple curve is the dose-response curve derived from the average inhibitory activities or pooled activities of geldanamycin at its corresponding concentrations. Y axis = geldanamycin inhibitory activity; x axis = geldanamycin concentration.
(D)Western blot shows 1uM of geldanamycin (GA) and its analog 17-DMAG block KDM4B activity in U2OS cells, as evidenced by changes in histone methyl marks assessed by specific antibodies, and inhibit the chaperone activity of HSP90 as evidenced by HSP70 compensatory upregulation.
(E) MALDI-FTICR mass spectrometry shows that KDM4B converts H3K9me3 to H3K9me2. Representative mass spectra of histone H3 peptide in the absence (upper panel) and presence (lower panel) of KDM4B. m/z = 1530.871 and 1516.854 peaks correspond to trimethylated H3K9 (substrate) and dimethylated H3K9 (product) peptides, respectively. Y axis indicates the signal intensity of H3K9me3 and H3K9me2 peaks. m/z represents mass divided by charge number and the x axis is expressed in units of m/z.
(F) Dose-response curve of geldanamycin shows the inhibition of KDM4B with IC50 of 1.24 μM, measured by MALDI-FTICR. KDM4B inhibition plot in the presence of geldanamycin. Value for Ki was calculated upon fitting the plot with Morrison equation. Data are shown as mean ± standard deviation (n = 3). Y axis indicates the turnover rate of H3K9me3 to H3K9me2, catalyzed by KDM4B. X axis indicates the concentration of geldanamycin.
(G) Microscale Thermophoresis assay of direct binding of KDM4B and geldanamycin. Y axis represents the fractions of KDM4B protein bound by geldanamycin. X axis represents geldanamycin concentration. Kd = dissociation constant.