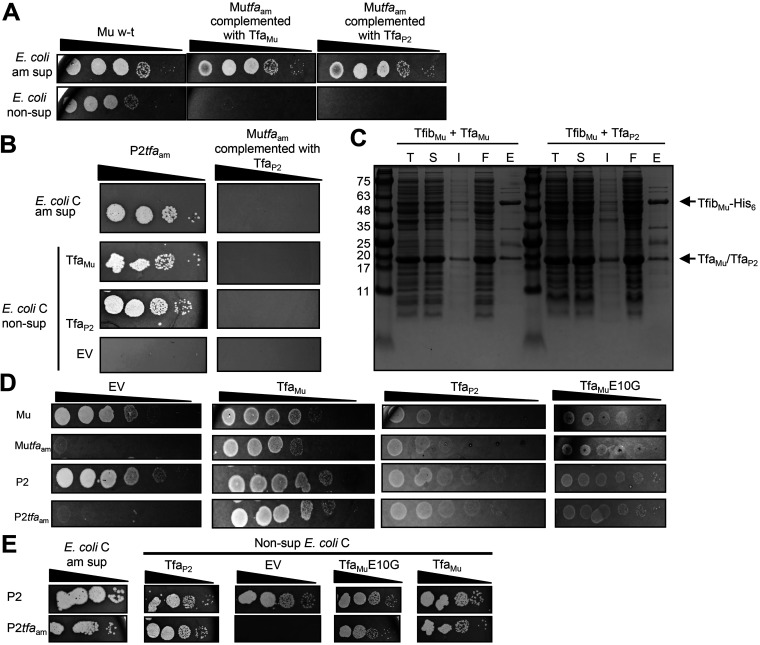
FIG 4.
Effects of Tfa expression on phage replication. (A) Serial 10-fold dilutions of lysates of Mu wild type, Mutfaam mutant phage complemented by TfaMu, or Mutfaam mutant phage complemented by TfaP2 were spotted on a lawn of E. coli K-12 amber suppressor and nonsuppressor cells. The images shown depict the zones of clearing produced on the bacterial lawns as a result of phage-mediated cell lysis. The infecting Mutfaam mutant particles contain TfaMu or TfaP2, but the zones of clearing observed on the suppressor strain are due to suppression of the amber mutation in subsequent rounds of phage growth. Although replication in these cells is due to suppression of the tfaam mutation, the initial infection of the cells would potentially be affected by the identity of the Tfa protein added by plasmid-based complementation. Data are representative of at least three biological replicates. (B) Serial 10-fold dilutions of lysates of P2tfaam, or Mutfaam complemented by TfaP2 were spotted on a lawn of E. coli C amber suppressor cells (top), or nonsuppressor cells transformed with an empty vector (EV, bottom), or a plasmid expressing either TfaMu or TfaP2 (middle). The infecting Mutfaam mutant particles contain TfaP2, the zones of clearing observed on the suppressor strain are due to suppression of the amber mutation in subsequent rounds of phage growth, and the zones of clearing observed on nonsuppressor strains result from complementation of the amber mutation with a plasmid-encoded TfaP2 or TfaMu. Although replication in these cells is due to suppression of the tfaam mutation, the initial infection of the cells would potentially be affected by the identity of the Tfa protein added by plasmid-based complementation. Data are representative of at least three biological replicates. (C) Cell lysates and fractions from protein purification experiments using Ni-NTA affinity chromatography were analyzed by SDS polyacrylamide gel electrophoresis stained by Coomassie blue. The indicated proteins were coexpressed and total cell lysate (T) and soluble (S) and insoluble (I) fractions of the lysate were assessed. The 6×-His-containing protein complexes were purified from the soluble fraction of the cell lysate. Samples from the eluted fraction from the purification (E) and the flowthrough fraction (F) are shown. (D) Serial 10-fold dilutions of Mu wild type, Mutfaam, P2 wild type, or P2tfaam phage lysate were spotted on a lawn of E. coli K-12 cells transformed with an empty vector (EV) or a plasmid expressing TfaMu, TfaP2, or TfaMuE10G. Data are representative of at least three biological replicates. (E) Serial 10-fold dilutions of P2 wild type or P2tfaam phage lysate were spotted on a lawn of E. coli C amber suppressor cells or nonsuppressor cells transformed with an empty vector (EV) or a plasmid expressing TfaP2, TfaMuE10G, or TfaMu. Data are representative of at least three biological replicates.