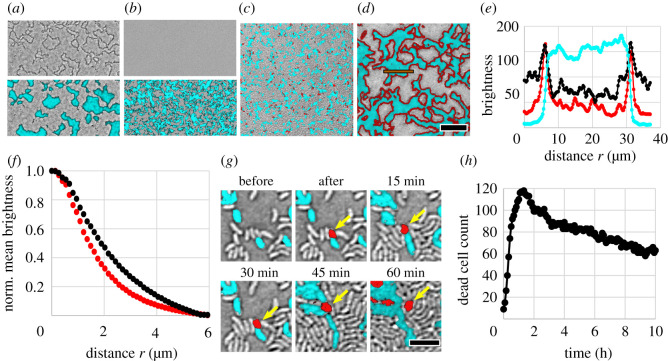
Figure 2.
Characterization of dead cell debris. (a) Representative bright-field image alone (top) and overlaid with fluorescence channel (bottom) of biofilms exhibiting unidirectional killing, recorded after 7 h of growth. Killer cells express superfolder green fluorescent protein (sfGFP) (cyan) while target cells are unlabelled. The entire field of view is densely packed with bacteria. (b) Same as (a) but the killer strain is engineered to be T6SS-deficient; no T6SS killing occurs and dark outlines are absent. (c,d) Merged bright-field and fluorescence images of unidirectional killing at start (0 h) (c) and after 7 h (d). Again, the killer expresses sfGFP (visible as cyan) while the target is unlabelled (grey). The DNA of compromised cells, i.e. dead cell debris, is labelled red via PI. Scale bar (a–d): 30 μm. (e) Intensity profile of different microscope images across a clonal patch (integrated vertically over the orange box in (d)) in the fluorescence images and bright-field image (inverted grey scale). (f) Normalized mean intensity of PI signal (red curve) and inverted bright-field signal (black curve) within a distance r from the interface between competing strains in (d). (g) Killing event, where a fluorescent killer cell kills a non-fluorescent target cell, which subsequently turns red. The dead cell debris is relocated by neighbouring cells that exert forces upon growth. Scale bar: 4 μm. (h) Counting the number of discrete PI-labelled cells over time demonstrates that PI-labelled dead cell debris persists long after cell death. Data were recorded from individual target cells densely surrounded by killer cells (initial target to killer number ratio of 1 : 50).