Figure 2.
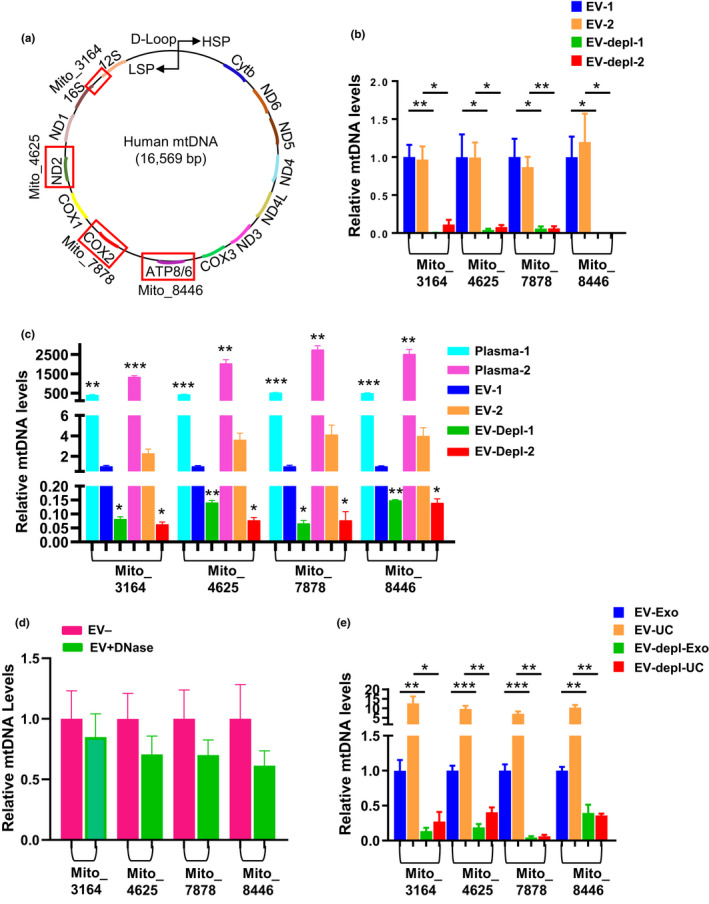
mtDNA is present in EVs (a) Schematic of mtDNA primer design. mtDNA primer regions are boxed in red and bolded next to each boxed region is the primer name and the starting nucleotide. Mito_3164 primer crosses the 16S rRNA and tRNA‐Ile1 regions and the forward sequence starts at bp 3164. Mito_4625 targets the ND2 region of the genome starting at bp 4625. Mito_7878 targets the COX2 region of the genome starting with bp 7878. Mito_8446 targets the ATP8 gene and begins at bp 8446. (b) Two sets of pooled plasma samples (from n = 4 individuals) were divided into four equal aliquots. EVs were isolated, and both EV and EV‐depleted fractions were used for DNA isolations. Four different primers were used to analyze mtDNA content in DNA derived from EV and EV‐depleted fractions using qPCR. (c) Plasma was pooled from the same individuals as in (b) and DNA was isolated from plasma, EVs or EV‐depleted fractions (n = 3 per condition). mtDNA was analyzed as above. (d) Plasma EVs were isolated with or without the DNase treatment step (n = 4 per condition). (e) Pooled plasma samples were divided into 8 equal aliquots. EVs were isolated for half of the aliquots using ultracentrifugation (UC) and the other half using ExoQuick™ (Exo). The EV and EV‐depleted (EV‐depl) fractions from each isolation method were used for DNA isolations for each histogram, and mtDNA levels were normalized to the EV group for each different primer set. Histograms represent the mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001 by Student t test
