Abstract
There are more than a thousand trillion specific synaptic connections in the human brain and over a million new specific connections are formed every second during the early years of life. The assembly of these staggeringly complex neuronal circuits requires specific cell-surface molecular tags to endow each neuron with a unique identity code to discriminate self from non-self. The clustered protocadherin (Pcdh) genes, which encode a tremendous diversity of cell-surface assemblies, are candidates for neuronal identity tags. We describe the adaptive evolution, genomic structure, and regulation of expression of the clustered Pcdhs. We specifically focus on the emerging 3-D architectural and biophysical mechanisms that generate an enormous number of diverse cell-surface Pcdhs as neural codes in the brain.
Keywords: Clustered protocadherins, Genome architecture, Neuronal identity, Adhesion specificity, Self-avoidance, Cell recognition
Introduction
The human brain contains a staggering 86 billion neurons, each with numerous branches of dendrites covering receptive fields and of axons innervating diverse regions with minimal overlap. The correct patterning of dendritic and axonal arbors is central for establishing and maintaining enormously complex networks with specific neuronal connectivity in the brain. These vast networks of synaptic connections between axons and dendrites form specific neuronal circuits to fulfill complicated cognitive functions and to determine personality traits and behavior. Aberrant assemblies of neuronal circuits underlie neuropsychiatric diseases. Neuronal circuit assemblies require each neuron to have an identity code for self-recognition and non-self discrimination. How these fascinating and diverse neuronal networks are generated is of the utmost importance. In addition, how the limited size of the human genome encodes the enormous number of neuronal cell-surface identity codes is intriguing.
Over the past few decades, great progress has been made to uncover large families of adhesion proteins that are candidates for cell-surface identity codes for neuronal circuit assembly, such as neurexins [1], olfactory receptors [2], cadherins and families of other adhesion molecules [3–6]. For example, in Drosophila melanogaster, 38,016 isoforms of Dscam1 (Down syndrome cell adhesion molecule 1)—generated by alternative splicing—endow each neuron with a unique identity code to discriminate self from non-self [7–10]. In vertebrates, this is achieved through the stochastic and combinatorial expression of ~60 clustered protocadherin (Pcdh) genes [11–13].
Cadherins are a superfamily of Ca2+-dependent cell-adhesion proteins that are required for specific cell-cell recognition in metazoans. Members of the cadherin superfamily include classical cadherins (type I and type II), clustered Pcdhs (α, β, γ), and non-clustered Pcdhs [6]. Compared with classical cadherins with five ectodomains (ECs), Pcdhs have six or more ECs with characteristic genome organization, in which multiple ECs are encoded by single unusually large exons [14, 15], and have diverse functions such as neuronal migration and axonal development [15, 16]. Clustered Pcdh genes are arranged in closely-linked clusters in one chromosomal region, while non-clustered Pcdh genes are scattered on different chromosomes [17]. As the largest subfamily of the cadherin superfamily, clustered Pcdh genes are prominently expressed in the brain, and each encodes a cadherin-like protein with six characteristic EC repeats. Their variable and constant genomic architectures are remarkably similar to those of the immunoglobulin (Ig), T cell receptor (Tcr), and UDP glucuronosyltransferase (Ugt) gene clusters, which generate tremendous diversity for the humoral immunity, cellular immunity, and chemical defense systems, respectively [11, 18].
In this review, we describe 3-D architectural and biophysical mechanisms for Pcdh neural codes in the brain. We first describe the 1-D genomic organization of the three Pcdh gene clusters and the 3-D architectural mechanisms that generate their combinatorial repertoires for single neurons. We then discuss cis- and trans-interactions between the extracellular domains of cell-surface Pcdh proteins to ensure neurons for self-recognition as well as self and non-self discrimination. These interactions transduce extracellular contact-dependent signals into the cytoplasm to induce actin dynamics and cytoskeletal remodeling through the common intracellular constant domains. It is this cytoskeletal remodeling that leads to the many functions of Pcdh such as neuronal migration, neurite morphogenesis, dendritic self-avoidance, axonal projection, spine elaboration, synaptogenesis, and neuronal connectivity. We refer interested readers to other excellent reviews discussing various aspects of the clustered Pcdh genes [5, 6, 19–25].
If It Looks Like a Code and Organizes Like a Code, It is a Code
Genetic studies have a long history of describing the phenomena of heredity. While individual genes determine certain phenotypes, the genome with the entire gene assembly holds the characteristics of a species and every creature has a genome that is passed on to the next generation. The genome encodes the brain, but the environment shapes and sharpens the brain: so-called neural epigenetics. The complexity of the brain determines the mind and consciousness. Both the brain and genome code and store information that is vital for the life of creatures. While the genome and genetic codes have been decoded [26, 27], the nature of the neural codes that wire the brain is still under intense investigation.
Setting the Stage for Neural Identity Codes
In the early 1940s, the Chemoaffinity Hypothesis posited that neurons express on their plasma membranes individual identification tags that specify synaptic connections [28]. Intensive efforts have since been devoted to uncovering the proposed neural codes but the exact nature of the neuronal chemoaffinity tags remains elusive [29, 30]. Among the four cell-adhesion families of cadherins, selectins, integrins, and Ig-containing proteins, cadherins are the only family that functions in direct Ca2+-dependent plasma membrane-to-membrane homotypic interactions, and are thus strong candidates for the chemoaffinity tags of neural codes in the brain [3, 5, 6, 31, 32]. However, only about a dozen classical cadherin genes and a few Pcdh genes were cloned in the nineties [33, 34]. Using the yeast two-hybrid system, 2 full-length and 6 partial cadherin-related receptor genes were cloned from mouse brain tissues and found to be expressed at synaptic junctions in neuronal subpopulations [35]. However, where exactly these proteins are located remains to be determined.
It turned out that these genes are members of the Pcdhα cluster which happens to be located upstream of the two other large gene clusters of Pcdhβ and Pcdhγ [11]. In total, there are 15 Pcdhα, 16 Pcdhβ, and 22 Pcdhγ genes that are highly similar and organized in tandem arrays in a single locus of the human genome. These large numbers and the striking organization immediately suggest that the clustered Pcdh genes are the long-sought neuronal address codes for the brain [4, 36–38]. These numbers are orders of magnitude less than that of neurons in the brain; however, mathematic analyses suggest that they are enough to encode the synaptic address codes required for geometrically constrained local brain regions or nuclei [39].
Genomic Organization of Clustered Pcdh Genes
The mammalian clustered Pcdh proteins are encoded by three closely-linked gene clusters (Pcdhα, Pcdhβ, and Pcdhγ) which span nearly 1 million base pairs [11]. The genomic arrangements of the Pcdhα and Pcdhγ clusters are similar, both with tandem arrays of large variable exons followed by respective single sets of three small constant exons (Fig. 1A) [11, 14, 40]. Within the Pcdhα and Pcdhγ clusters, each variable exon carries its own promoter and can be spliced to the single set of downstream constant exons of its respective cluster. Through stochastic promoter activation and cis-alternative splicing, clustered Pcdhs can generate dozens of different isoforms [41, 42].
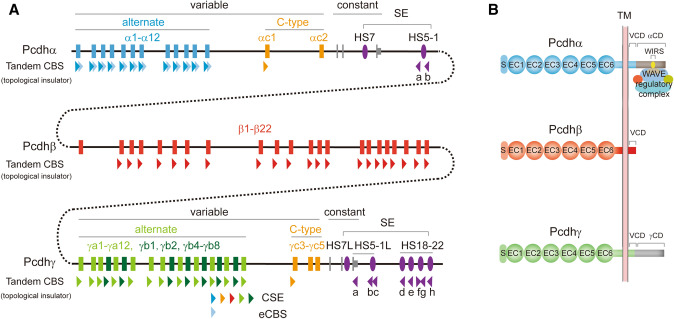
Fig. 1.
Genomic organization and domain structure of clustered protocadherins. A Mouse clustered protocadherin genes have 58 isoforms arranged into three closely-linked clusters: Pcdh α, β, and γ. The Pcdh α and γ gene clusters contain more than a dozen of unusually large, highly similar, and repetitive variable exons, each of which is associated with a promoter and can be spliced to a common set of three downstream small constant exons within the respective cluster. These variable exons can be separated into alternate and C-type groups, based on the encoded protein sequence similarity. The Pcdhβ gene cluster lacks constant exons and only contains 22 variable exons each of which encodes a full-length protein. HS7 and HS5-1 constitute a super-enhancer (SE) for the Pcdhα cluster. HS7L, HS5-1L, and HS18-22 constitute a super-enhancer for the Pcdh β and γ clusters. The locations and relative orientations of tandem CTCF sites (CBS, CTCF binding site), which function as topological insulators, are marked as arrowheads under the respective promoters and enhancers. Note that each Pcdhα alternate promoter is flanked by two CBS elements (CSE and eCBS). HS, DNaseI hypersensitive site. B The domain organization of the encoded protein structure of clustered Pcdhs. Each large variable exon encodes an extracellular domain with a signal peptide, followed by 6 ectodomain (EC) repeats, a transmembrane (TM) domain, and a juxtamembrane variable cytoplasmic domain (VCD). The three small constant exons encode a common membrane-distal intracellular constant domain (CD) shared by all isoforms of the Pcdh α or γ cluster. There is a WAVE interacting receptor sequence (WIRS) motif located near the C-terminal end of the Pcdhα CD that recruits the WAVE-regulatory complex and links to actin cytoskeletal dynamics.
The variable exons of Pcdhα and Pcdhγ can be further divided into alternate and C-type gene groups based on their genomic location and sequence similarity (Fig. 1A). The mouse Pcdhα cluster contains 12 alternate genes (α1–α12) and two C-type genes (αc1 and αc2). The mouse Pcdhγ cluster contains 19 alternate genes (12 A-types: γa1–γa12; 7 B-types: γb1, γb2, γb4–γb8) and three C-type genes (γc3–γc5). Different from Pcdhα and Pcdhγ, the mouse Pcdhβ cluster, however, contains 22 genes (β1–β22) and no C-type gene (Fig. 1A). In total, there are five C-type variable exons that are more similar to each other than to members of the alternate gene group [11, 40]. However, Pcdhβ contains only large variable exons and lacks constant exons (Fig. 1A). Therefore, each member of the Pcdhβ cluster is a single-exon gene [11, 40]. Together, these three clusters encode 58 Pcdh isoforms (14α, 22β, and 22γ) in mice and 53 Pcdh isoforms (15α, 16β, and 22γ) in humans (Fig. 1A).
In the Pcdhα cluster, the promoter of each alternate gene is flanked by two CTCF-binding sites (CBS or CTCF sites). In the Pcdhβ cluster, the promoter of each gene is associated with one CBS element except β1 which has no CBS element (Fig. 1A). In the Pcdhγ cluster, the promoter of each alternate gene is associated with one CBS element. Finally, among the five C-type Pcdh genes, only the first C-type gene of the Pcdhα cluster (αc1) and the first C-type gene of the Pcdhγ cluster (γc3) are associated with a CBS element (Fig. 1A).
Each variable exon encodes a signal peptide, followed by an extracellular domain containing 6 ECs, a transmembrane region, and a juxtamembrane variable cytoplasmic domain (VCD). The three constant exons encode a common membrane-distal intracellular constant domain (CD) shared by all members of the Pcdhα or Pcdhγ family. Since the Pcdhβ cluster has only a variable region with no constant region, each Pcdhβ variable exon is an independent gene, which encodes a Pcdh protein with an extracellular domain of 6 ECs, a transmembrane region, and a short VCD, but lacks a common CD (Fig. 1B) [11, 14].
The Pcdha cluster is regulated by a super-enhancer composed of two cis-regulatory elements, HS7 and HS5-1 (HS, hypersensitive site) (Fig. 1A). Similarly, a super-enhancer, composed of HS7L (HS7 like), HS5-1L (HS5-1 like), and HS18-22, was also identified downstream of the Pcdhγ cluster for both the Pcdhβ and Pcdhγ clusters (Fig. 1A) [43–48].
Fifteen DNaseI hypersensitive sites (HS15-HS1) were initially identified in the Pcdhα cluster, among which HS7 and HS5-1 have strong enhancer activity in a transgenic reporter assay [49]. In mice, genetic deletion of HS5-1, which is located 30 kb downstream of the last Pcdhα constant exon, results in a significant decrease in the expression levels of Pcdhα1–α12 and Pcdhαc1 in the brain, but does not affect the expression of Pcdhαc2 [48, 50]. By contrast, deletion of HS7, which is located between the constant exons 2 and 3, results in a significant decrease of expression levels of all Pcdhα genes, including Pcdhαc2 [50].
Adaptive Evolution of Clustered Pcdh Genes
Initial studies on Pcdh genes showed that the encoded extracellular domain contains a “primordial” cadherin motif, similar to cadherin motifs in the Drosophila Fat protein [34]. It was thought that Pcdhs may be evolutionarily more ancient than the classical cadherins [34]. In addition, the Pcdh genes have characteristic genomic organizations in which multiple ECs are encoded by large exons, a feature that is distinct from the genomic organizations of classical cadherins [14]. Complete sequencing of the Drosophila genome revealed, however, that it does not contain clustered Pcdh genes [51]. Thus, the “proto” affix in the “protocadherin” nomenclature is a misnomer and the clustered Pcdh genes are thought to have adaptively evolved later and may be related to functions of more advanced nervous systems.
Similar to the human genome, the chimpanzee, mouse, and rat genomes contain the three Pcdh gene clusters [40, 52, 53]. Clustered Pcdh genes also exist in the anole lizard, frog, coelacanth, fugu, and zebrafish [52, 54–59]. The genome of the frog Xenopus tropicalis contains the Pcdh α and γ clusters but lacks Pcdhβ; however, the Pcdhγ cluster has been duplicated into two clusters [59]. In addition, the fugu and zebrafish genomes lack the Pcdhβ cluster but contain two Pcdh α and γ clusters because of the whole-genome duplication in the ray-finned lineage [52, 54, 57].
The anole and coelacanth genomes contain the Pcdhβ cluster [55, 58]. This suggests that the Pcdhβ cluster in mammals, anole, and coelacanth probably results from the duplication of variable exons of the Pcdhγ cluster. The duplicated variable exons subsequently lost their ability to be spliced to the constant exon 1 of the Pcdhγ cluster. Another possibility is that the Pcdhβ cluster results from duplication of the entire Pcdhγ cluster. The duplicated cluster then lost its constant exons through mutation or degeneration. Further research is needed to distinguish these two scenarios. Nevertheless, the topological regulation of both Pcdh β and γ clusters by a single super-enhancer composed of tandem arrays of CTCF sites (Fig. 1A) suggests that they share a common ancestor [48], consistent with their evolutionary trees [52]. Finally, molecular and structural analyses revealed that Pcdhβ and Pcdhγ share characteristics that are distinct from Pcdhα [12, 60].
The cartilaginous shark genome contains a single locus composed of four closely-linked Pcdh clusters that are para-orthologous to the three mammalian Pcdh gene clusters, suggesting that the ancestral jawed vertebrates contained seven Pcdh gene clusters [61]. During the evolution of the genomes of cartilaginous fish and bony vertebrates, this ancestral Pcdh locus experienced differential losses in that the mammalian lineages lost four clusters and the shark lineage lost three clusters [61]. Interestingly, clustered Pcdh genes are vastly expanded in the invertebrate octopus genome and enriched in neural tissues, consistent with their roles in establishing and maintaining the large and complex octopus nervous system [62, 63].
3-D Genome Architecture of Clustered Pcdhs
The three Pcdh gene clusters are organized as a large superTAD (super topologically associating domain) which can be divided into two subTADs of α and βγ (Fig. 2A) [45, 64]. The Pcdhα subTAD is formed by long-distance chromatin interactions between tandem arrays of forward CBS elements or CTCF sites of the variable region and the two reverse CBS elements flanking HS5-1 (Fig. 2A). The Pcdhβγ subTAD is formed by long-distance chromatin interactions between tandem arrays of forward and reverse CBS elements within the promoter and super-enhancer regions (Fig. 2A). We outline the important role of higher-order chromatin structures in the regulation of clustered Pcdhs in this section.
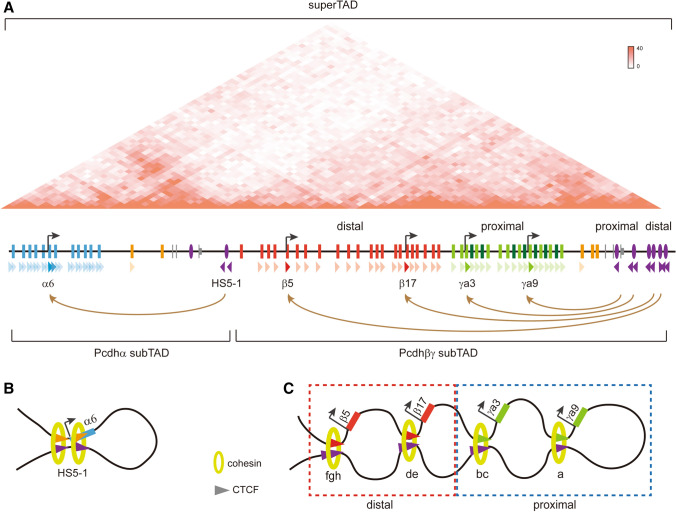
Fig. 2.
Topological regulation of the clustered Pcdh genes in single alleles by tandem CTCF sites. A Hi-C map showing the three Pcdh clusters are organized into one superTAD composed of two subTADs. In the Pcdhα cluster, each promoter-associated CTCF site functions as a topological insulator for all of its upstream genes. In the Pcdhβγ clusters, tandem CTCF sites function as topological insulators, resulting in proximal-proximal and distal-distal CBS interactions. B HS5-1 forms spatial chromatin contacts with one and only one chosen alternate promoter through CTCF/cohesin-mediated “double-clamp” looping in the Pcdhα cluster. C Tandem CTCF sites function as topological insulators to balance spatial chromatin contacts and enhancer-promoter selection. The proximal CTCF sites of the super-enhancer form long-distance chromatin interactions with the Pcdhγ cluster while the distal CTCF sites of the super-enhancer form long-distance chromatin interactions with the Pcdhβ cluster, reminiscent of nested cohesin-extruded loops in the extended “Hulu model” [48].
CTCF Protein as a Key 3-D Chromatin Architect
CTCF (CCCTC-binding factor) is the best-characterized insulator-binding protein in mammals that organizes the 3-D architecture of the genome. It regulates gene expression of the clustered Pcdhs through mediating long-range chromatin contacts between remote enhancers and target promoters. The topological chromatin loops between enhancers and promoters are formed by cohesin-mediated active loop extrusion [48]. Cohesin, a ring-shaped complex embracing double-stranded DNA, continuously extrudes chromatin fibers until blocked by CTCF-bound CBS elements. The cohesin loop extrusion brings the two remote DNA fragments with forward-reverse convergent CBS elements into close contact in the 3-D nuclear space [45, 65–67].
There is substantial evidence for a central role of CTCF/cohesin in clustered Pcdh gene regulation. First, knockdown of cohesin results in the loss of chromatin loops and downregulation of the clustered Pcdh genes [44, 68]. Second, knockdown of CTCF in cell lines also results in the loss of chromatin loops and a significant decrease of Pcdh expression levels [44, 68, 69]. Finally, conditional knockout of CTCF in neurons markedly downregulates a staggering 53 out of the total 58 clustered Pcdh genes in mice [70], providing strong evidence that CTCF is a master regulator for clustered Pcdhs [69].
Oriented CTCF Sites as Codes of Articulation Joints for Building 3-D Genome Architecture
Initial computational analyses identified a conserved sequence element (CSE), with a highly-conserved CGCT box, located at about the same distance upstream of the translational start codon of each member of the three Pcdh clusters (except for αc2, β1, γc4, and γc5) [40]. These CSEs were later shown to bind CTCF proteins, and thus are CBS elements (Fig. 1A) [44, 68, 69].
In the Pcdhα cluster, there is an additional CBS element located at ~700 bp downstream of the CSE within the coding region of each alternate variable exon (known as eCBS for exonic CBS) (Fig. 1A) [44, 68]. Thus, there are two CBS elements (CSE and eCBS) flanking each Pcdhα alternate promoter. However, there is only one CBS element (CSE) associated with the αc1 promoter and no CBS element associated with the αc2 promoter (Fig. 1A). Interestingly, the HS5-1 enhancer is also flanked by two CBS elements, HS5-1a and HS5-1b, with an intervening distance similar to that between each promoter-flanking CBS pair of CSE and eCBS (Fig. 1A) [44, 68].
All of the CBS elements (CSE and eCBS) flanking the Pcdhα promoters are in the forward orientation. By contrast, the two CBS elements (HS5-1a and HS5-1b) flanking the HS5-1 enhancer are in the reverse orientation (Fig. 1A). Namely, the CBS elements in the Pcdhα promoter and enhancer regions are in the opposite orientation [44]. Forward-oriented CBS elements flanking a Pcdhα promoter and reverse-oriented CBS elements flanking the HS5-1 enhancer interact spatially to form a “double-clamp” transcription hub through CTCF/cohesin-mediated chromatin looping (Fig. 2B) [44, 71].
CTCF Site Orientation Determines the Directionality of Chromatin Looping
Inversion of the two enhancer CBS elements (HS5-1a and HS5-1b) in cells and mice by using the CRISPR/Cas9-mediated DNA fragment editing method provides strong evidence for the causality between CBS orientation and chromatin-looping directionality [45, 48]. Specifically, the reverse-oriented CBS elements flanking the HS5-1 enhancer normally form long-distance chromatin interactions with the forward-oriented CBS elements associated with the upstream Pcdhα promoters (Fig. 2A, B) [44]. After inversion by CRISPR DNA-fragment editing [72, 73], however, they no longer form long-distance chromatin interactions with the upstream Pcdhα promoters. Strikingly, the inverted CBS elements form long-distance chromatin interactions with the downstream CBS elements [45]. Thus, the relative orientation determines the directionality of long-distance chromatin looping [45]. In addition, spatial chromatin contacts are preferentially formed between forward-reverse CBS elements through CTCF/cohesin-mediated loop extrusion throughout the entire genome [45, 47, 48, 65, 67, 74]. Finally, these experiments also provide strong in vivo evidence that enhancers do not function in an orientation-independent manner, at least those associated with CBS [45].
Tandem CTCF Sites as Genome Topological Insulators
In the Pcdhβγ clusters, only a single CTCF site is associated with each variable promoter (except β1, γc4, and γc5) (Fig. 1A) [44, 68, 70]. Similar to the Pcdhα cluster, all of the promoter CTCF sites are in the forward orientation in the Pcdhβγ clusters. By contrast, the downstream super-enhancer contains several reverse-oriented CTCF sites organized in tandem (Fig. 1A) [45, 46, 48].
Genetic deletion of HS18-20 (part of the super-enhancer [46]) in mice results in a significant decrease of expression levels of the Pcdhβ genes [43]. In addition, deletion or inversion of HS5-1bL together with HS18-20 in mice totally abolishes the expression of all Pcdhβ genes, suggesting that these regulatory elements, bypassing the Pcdhγ cluster, are enhancers for members of the Pcdhβ cluster [43, 47]. However, the expression levels of the Pcdhγ genes are mostly unaffected in these deletions, leaving the regulation of the Pcdhγ genes an unresolved question [43, 47].
The Pcdhβγ genes are topologically regulated by the tandem CTCF sites of the downstream super-enhancer. Specifically, chromosome conformation capture experiments have revealed that the Pcdhγ genes are in close spatial contact with the proximal CTCF sites of the super-enhancer (Fig. 2A, C). By contrast, the Pcdhβ genes are in close spatial contact with the distal CTCF sites of the super-enhancer (Fig. 2A, C) [48]. This topological regulation solves the long-standing mystery of Pcdhγ gene regulation.
These proximal-to-proximal and distal-to-distal topological chromatin regulations were further confirmed by a series of genetic manipulations of the CTCF sites in the super-enhancer in vivo. Specifically, when CTCF sites in the super-enhancer are deleted or inverted, the downstream reverse-oriented CTCF sites show increased chromatin interactions with members of the Pcdhγ cluster and decreased chromatin interactions with members of the Pcdhβ cluster [47, 48]. Thus, tandem CTCF sites function as topological insulators to mitigate the chromatin contacts with and usage of the proximal Pcdhγ promoters. In addition, these topological insulators, counter-intuitively, promote chromatin contacts with and the usage of the distal Pcdhβ promoters. In conclusion, tandem arrays of oriented CBS elements determine the allocation of spatial resources of enhancers for promoters of both distal and proximal Pcdh genes.
Epigenetic Regulation of Chromatin Loops
Methylation of the CpG dinucleotide within the CGCT box of the CTCF sites of Pcdh promoters precludes CTCF binding, suggesting epigenetic regulations of the clustered Pcdh genes [44]. In each cell, these CTCF sites are differentially methylated, with one and only one alternate exon being activated through long-range chromatin contacts with the HS5-1 enhancer (Fig. 2B) [48, 75–77]. In the neuroblastoma cell line SK-N-SH, Pcdhα expression levels are inversely correlated with promoter methylation. Specifically, the CBS elements of expressed isoforms are unmethylated and bound by CTCF, but the CBS elements of silenced isoforms are methylated and devoid of CTCF proteins [44]. Consistently, demethylation of CBS elements activates Pcdhα gene expression [78]. Finally, recent structural analyses suggest that the addition of a methyl group at the 5th position of cytosine within the CpG interferes with the binding of CTCF zinc finger 3 to the CGCT box [79].
In neurons, the DNA methylation states of the Pcdh promoters are also inversely correlated with the transcription states of the Pcdh genes. For example, alternate Pcdhα genes, which are stochastically expressed by individual Purkinje cells, show mosaic and differential methylation patterns. In contrast, the C-type isoforms, which are constitutively expressed, are hypomethylated [75]. Thus, stochastic expression of Pcdh isoforms is probably determined by the DNA methylation in individual neurons.
Recent studies revealed that the eCBS element of each alternate exon is associated with an antisense promoter which transcribes a long non-coding RNA (lncRNA) [78]. Stochastic transcription of this lncRNA extends through the sense promoter, leading to DNA demethylation of the corresponding CBS element. This CBS demethylation then facilitates CTCF binding and subsequent activation of the sense promoter [78]. Interestingly, the promoter activation mediated by antisense lncRNA transcription is only found in alternate but not C-type Pcdhα genes. This is consistent with the fact that the C-type Pcdhα variable exons do not contain an eCBS element.
Other Potential Regulatory Proteins
In addition to the architectural proteins CTCF and cohesin, other potential 3-D genome architectural proteins have been shown to regulate expression of the clustered Pcdh genes. For example, a protein known as structural maintenance of chromosome hinge domain containing 1 (SMCHD1), which is critically involved in the pathogenesis of facioscapulohumeral muscular dystrophy, antagonizes CTCF in Pcdh gene regulation [80]. The SMCHD1 occupancy at Pcdhα promoters and enhancers coincides with CTCF sites. Loss of Smchd1 results in increased CTCF binding to the Pcdhα alternate promoters and upregulation of Pcdh α and β gene expression [80]. However, the underlying mechanism by which SMCHD1 antagonizes CTCF DNA binding remains unknown.
SET domain bifurcated 1 (Setdb1) is required for the maintenance of the superTAD structure in Pcdh clusters [64]. Conditional knockout of Setdb1 in forebrain neurons results in the loss of H3K9me3, leading to demethylation of DNA and subsequent recruitment of CTCF to Pcdh promoters [64]. The increased CTCF binding strengthens the chromatin interactions between Pcdh promoters and enhancers, but weakens the chromatin interactions between the boundaries of the superTAD. Neurons without Setdb1 lose the stochastic constraint and express increased numbers of Pcdh isoforms [64].
Neuron-restrictive silencer factor (NRSF) regulates the neuron-restrictive expression of Pcdhα through binding to HS5-1 and Pcdhα variable exons [50, 81]. In addition, Wiz (widely-interspaced zinc finger-containing protein) defines cell identity by functioning as a DNA loop anchor in collaboration with CTCF and cohesin [82]. Wiz has been shown to regulate Pcdhβ gene expression in mice [83]. Consistently, Wiz proteins are enriched at all of the Pcdhβ promoters (except Pcdhβ1, which is the only Pcdhβ gene with no CTCF site) and at the HS5-1bL site of the Pcdhβγ super-enhancer [83]. All in all, various transcription factors may regulate the stochastic expression of clustered Pcdhs by altering higher-order architectural chromatin loops between enhancers and promoters.
Mechanisms for Generating Clustered Pcdh Codes of Neuronal Identity
Combinatorial Expression of Pcdhs as Cell-Surface Identity Codes
Each cortical neuron stochastically expresses up to 2 alternate Pcdhα genes, 4 Pcdhβ genes, and 4 alternate Pcdhγ genes as well as all of the 5 C-type Pcdh genes (up to 15 in total) [48, 84]. These combinatorial expression patterns could generate the large number of address codes required for neuronal identity. For example, the 22 encoded Pcdhγ proteins have been predicted to form up to 234,256 distinct tetramers of cell-surface assemblies [85]. In conjunction with the encoded 15 Pcdhα and 22 Pcdhβ proteins, Pcdh proteins could generate the enormous diversity of cell-surface assemblies required for coding single neurons in the brain. We summarize the mechanisms of Pcdh promoter choice and expression regulation in this section.
Establishment and Maintenance of Clustered Pcdh Expression Patterns
A remarkable property of the clustered Pcdh genes is that their promoter choice is inherited and stably maintained by daughter cells as seen in the SK-N-SH cell line and differentiated neurons [44, 86]. This suggests that, once chosen, the expression patterns of clustered Pcdh genes are epigenetically inheritable. In addition, Pcdh promoter choice occurs early during the naive-to-primed conversion of ESCs (embryonic stem cells) [86]. The Pcdh promoters are modified with both active (H3K4me3) and repressive (H3K27me3) chromatin marks, so called bivalent promoters, in the primed ESCs before being activated. The chosen Pcdh genes are then stably inherited by differentiated neurons [86].
As the methylation states of promoters are inversely correlated with the expression levels of clustered Pcdh genes, a fundamental question is how single neurons achieve the stochastic activation of Pcdh promoters. On the one hand, stochastic activation of a Pcdh promoter could be achieved through demethylation of the chosen target promoter by antisense transcription of lncRNA [78]. On the other hand, this could be achieved through methylation of all of the non-chosen promoters [75]. Consistently, all of the Pcdhα alternate promoters are enriched with CTCF in naive ESCs, while only chosen promoters are enriched with CTCF in primed ESCs [86], suggesting hypomethylation-to-hypermethylation conversion of the non-chosen promoters during cellular differentiation. This indicates that the ground state of Pcdh promoters is unmethylated or hypomethylated and that the activation of specific promoters requires methylation of all of the other promoters (Fig. 3A, B).
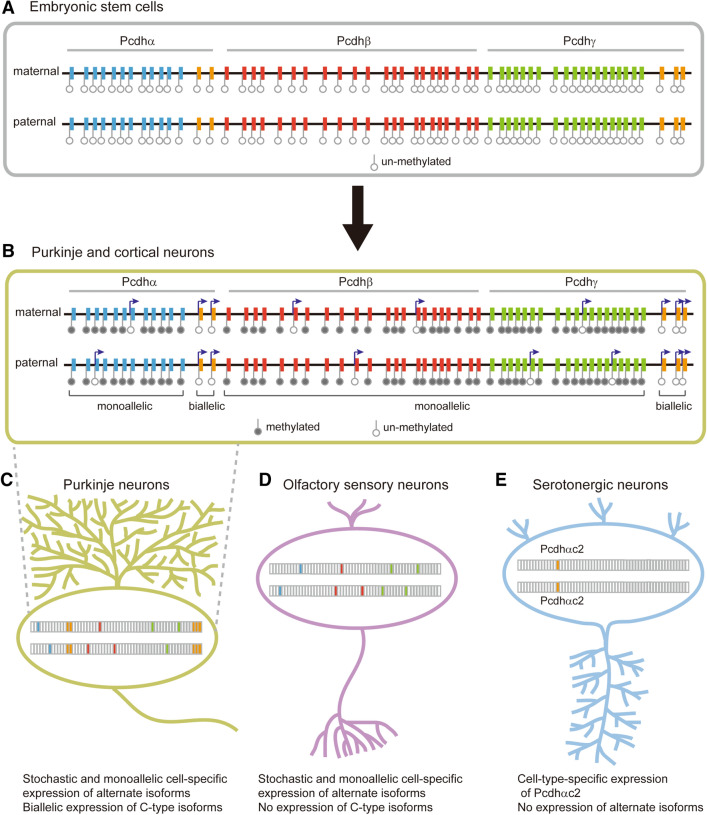
Fig. 3.
Cell-specific and cell-type-specific expression of clustered Pcdh genes. A In embryonic stem cells, the ground state of clustered Pcdh gene promoters is unmethylated. B Gene regulation of the three Pcdh clusters by DNA methylation in mature neurons such as Purkinje and cortical cells. C In the Purkinje neurons of the cerebellum, alternate genes are stochastically and monoallelically expressed in a cell-specific manner, while C-type genes are constitutively and biallelically expressed presumably in every cell. D In olfactory sensory neurons, alternate genes are stochastically and monoallelically expressed in a cell-specific manner, while the C-type genes are not expressed. E In serotonergic neurons in the raphe nuclei of the midbrain, only Pcdhαc2 is expressed in a cell-type-specific manner.
Cell-Specific and Stochastic Expression of Clustered Pcdh Genes
Clustered Pcdhs are widely expressed in the developing and adult central nervous systems [11, 34, 35, 42, 53, 87–90]. The expression of members of the Pcdhα cluster is highly specific to the central nervous system. While members of the Pcdh β and γ clusters are prominently expressed in the central nervous system, they are also expressed in several other tissues such as the kidney and lung [87, 89, 91]. Detailed expression patterns of each isoform were initially analyzed by in situ hybridization using isoform-specific probes, which showed that they are stochastically expressed in neuronal subpopulations in various brain nuclei or regions [35, 42, 53, 89, 90, 92].
Single Purkinje neurons express alternate members of clustered Pcdh genes in a stochastic and monoallelic manner (Fig. 3C) [92–94]. In addition, single cortical neurons also express alternate members of the three Pcdh clusters in a similar manner [48, 75, 84]. In the Pcdhα cluster, each tandem pair of the promoter CTCF sites (CSE and eCBS) functions as an insulator for all of its upstream Pcdhα genes. A single chromatin loop between the HS5-1 enhancer and a variable promoter determines the expression of the chosen Pcdhα gene in each allele (Fig. 2A, B) [48].
In the Pcdhβγ clusters, the super-enhancer is composed of four CBS-containing elements. Up to two Pcdhβ genes (activated by enhancers with CTCF sites “de” and “fgh”) and two alternate Pcdhγ genes (activated by enhancers with CTCF sites “a” and “bc”) could be expressed from each allele through nested chromatin loops (Fig. 2A, C) [48].
In olfactory sensory neurons (OSNs), clustered Pcdh genes are stochastically expressed, except for the C-types (Fig. 3D) [76]. In addition, diploid chromatin conformation capture of single OSNs has shown that there are significant cell-to-cell heterogeneities of Pcdh chromatin architectures and that Pcdh enhancers communicate with distinct Pcdh promoters in different cells [95]. This may reflect the stochastic Pcdh promoter choice. Specifically, each OSN expresses a distinct set of up to 10 alternate Pcdh genes, among which 5 are stochastically and monoallelically expressed from each allele (Fig. 2). In summary, these findings suggest that the clustered Pcdh genes are stochastically expressed in single neurons of the cerebellum, cerebrum, and olfactory epithelium in a cell-specific manner.
Cell Type-Specific Expression of Clustered Pcdh Genes
All of the C-type Pcdh genes appear to be constitutively and biallelically expressed in single neurons of the cerebellum and cerebrum in the mouse brain (Fig. 3B, C) [48, 75, 84, 92–94]. By contrast, none of the C-type Pcdh genes is expressed in mouse OSNs (Fig. 3D) [76]. Finally, only Pcdhαc2 is predominantly expressed in serotonergic neurons (Fig. 3E) [96, 97]. Collectively, these studies suggest that C-type Pcdh genes are expressed in a cell-type-specific manner, in stark contrast to the stochastic expression of alternate Pcdh genes in the brain.
Molecular Logic of Neuronal Self-avoidance and Coexistence
Promiscuous Cis-interactions for Diverse Cell-Surface Assemblies
The Pcdhα proteins co-immunoprecipitate with Pcdhγ in cell lysates. In addition, cell-surface delivery of Pcdhα proteins (except for Pcdhαc2) requires the co-expression of Pcdhγ because Pcdhα alone cannot be sufficiently expressed at the plasma membrane [12, 98], suggesting that Pcdhα and Pcdhγ may form heterodimers. Moreover, distinct members interact with each other in membrane fractions [85, 99]. Finally, each member of Pcdhβ or Pcdhγ (except for Pcdhγc4) can form homodimers or heterodimers; however, members of Pcdhα and Pcdhγc4 cannot form homodimers. They can only form heterodimers with Pcdhβ or other Pcdhγ isoforms [12, 100].
Structural studies support the formation of cis-homodimers or cis-heterodimers between isoforms of clustered Pcdhs. The cis-dimerization requires both EC5 and EC6 domains [13, 60, 101]. Specifically, the Pcdh cis-dimer interfaces are asymmetric, with one molecule providing the EC5 and EC6 side of the interface, and the other providing only the EC6 side (Fig. 4A) [13, 60]. Isoforms of Pcdhβ and Pcdhγ (except for Pcdhγc4) form cis-homodimers or cis-heterodimers in that each isoform can participate as either the EC5–EC6 or EC6 side of the interface [13, 60]. However, isoforms of Pcdhα and Pcdhγc4 can only form cis-heterodimers and cannot form cis-homodimers because they cannot participate as the EC6 side of the interface. Namely, they participate only as the EC5–EC6 side of the heterodimer interface. They need isoforms of either Pcdhβ or Pcdhγ (also known as carrier isoforms, except for Pcdhγc4) to provide the EC6 side of the heterodimer interface [60].
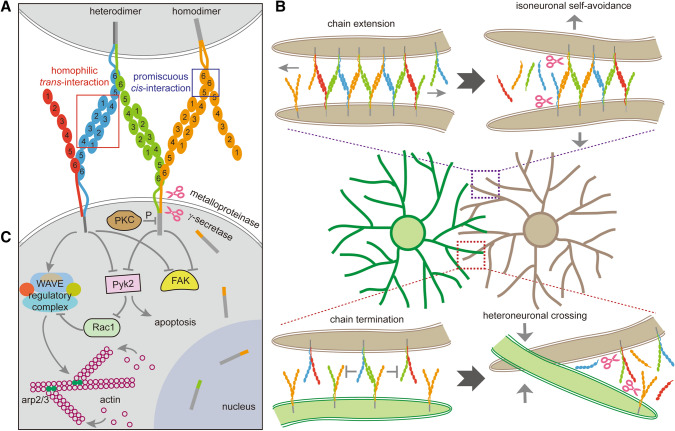
Fig. 4.
The molecular basis of self-recognition for self-avoidance and non-self coexistence mediated by clustered Pcdhs. A Clustered Pcdh isoforms form heterodimers and homodimers in the cell membrane through promiscuous cis-interactions between the EC5–EC6 domains of one isoform and the EC6 domain of the other isoform to endow each cell with an identity code. Clustered Pcdh isoforms from different neurites recognize each other through strict homophilic trans-interactions of the EC1–EC4 domains in an anti-parallel fashion. B Molecular arrangements of an extended self-recognition complex between identical combinatorial profiles expressed on the same neuron, resulting in adhesion-mediated repulsion between sister neurites from single neurons (isoneuronal self-avoidance). Specifically, when the expressed isoforms are the same between two neurites (from the same cell, for example), Pcdh isoforms linearly assemble into parallel arrays through cis- and trans-interactions to form larger zipper-like lattices between membranes. These structures trigger intracellular signaling and cytoskeletal rearrangement. Subsequent Pcdh cleavage may result in neurite self-avoidance. By contrast, when the two neurites from different neurons stochastically express distinct combinations of Pcdh isoforms, their assembly is interrupted by the mismatched isoforms, as proposed by the isoform-mismatch chain-termination model. This results in heteroneuronal crossing and coexistence. C Intracellular signaling of the clustered Pcdhs. Pcdh isoforms are cleaved by metalloproteinase and γ-secretase into an extracellular fragment and an intracellular fragment. The latter may translocate into the nucleus to regulate gene transcription. PKC phosphorylates the intracellular domain of Pcdhγ. Isoforms of Pcdh α and γ clusters bind and inhibit the activities of Pyk2 and FAK through the Pcdh intracellular domain. The intracellular domain of Pcdhα isoforms recruits the WAVE complex through the WIRS motif and activates actin-filament branching. Pyk2 also inhibits Rac1 and disinhibits the WAVE complex. These intracellular signaling pathways eventually lead to cytoskeletal remodeling and sister-neurite repulsion.
In summary, clustered Pcdh isoforms appear as a cell-surface repertoire composed of homodimers and promiscuous heterodimers of members of all three Pcdh clusters on the plasma membrane of single neurons [12, 13, 60, 85, 100].
Homophilic Trans-interactions for Self-recognition
Great progress has been made in deciphering the trans-interactions of clustered Pcdh proteins for generating cell-recognition specificity. The trans-interactions of the Pcdh isoforms have been tested using an efficient cell-aggregation assay by transfecting two cell populations [12, 85, 101]. Different cell populations expressing the same combinations of Pcdh isoforms display strict homophilic interactions and can form cell aggregates, but those expressing different combinations of Pcdh isoforms cannot [12].
All of the clustered Pcdh β and γ isoforms, except for Pcdhγc4, can engage in robust and highly specific trans-homophilic interactions in cell aggregation assays. These isoforms are delivered to cell membrane, probably because they can form cis-homodimers [60]. Pcdhα (except for Pcdhαc2) and Pcdhγc4, on the other hand, cannot form cis-homodimers and cannot be delivered to cell membrane by themselves. Therefore, they cannot induce cell aggregates [12]. Pcdhαc2, however, is unique in that it can induce cell aggregates by itself because it can form cis-homodimers and be delivered to cell membrane [12].
The Pcdhα proteins can form cis-heterodimers with isoforms of Pcdh β and γ (except for Pcdhγc4). They can be delivered to cell membrane when they are co-expressed with Pcdh β or γ isoforms. Therefore, Pcdhα (except for Pcdhαc1) does induce cell aggregates through homophilic trans-interactions when co-expressed with Pcdh β and γ isoforms (except for Pcdhγc4). Finally, homophilic interactions are abolished when there is a single mismatched isoform between the two transfected cell populations [12].
Structural analyses revealed that the trans-homophilic interactions are mediated by EC1–EC4 in an antiparallel manner. These trans-interactions form a zipper-like ribbon structure in apposed plasma membranes. Specifically, the EC1, EC2, EC3, and EC4 of one isoform at a cell surface interact with the EC4, EC3, EC2, and EC1 of the same isoform from the apposed cell surface, respectively [13, 101–105]. Among the six EC domains of clustered Pcdhs, EC2 and EC3 have been positively selected for diversity during evolution and are thus the most diversified ECs in amino-acid residues [52]. Consistently, they determine the stringent specificity of trans-homophilic interactions [12, 85, 104].
The Chain-Termination Model for Non-self Discrimination
The crystal structure of the full-length extracellular domain of Pcdhγb4 reveals a zipper-like lattice through cis-interactions mediated by EC5–EC6/EC6 and trans-interactions mediated by EC1–EC4 [13]. When tethered to liposomes, Pcdh extracellular domains spontaneously assemble into zipper-like linear arrays through trans-homophilic interactions between Pcdh dimers [13]. These linear assemblies extend through the contacted membranes as a chain to form a larger lattice (Fig. 4B). In this chain termination model, once a certain size threshold is reached, the assemblies trigger intracellular Pcdh signaling pathways to regulate various cellular behaviors such as repulsion. By contrast, when mismatched isoforms are incorporated, the Pcdh chain extension terminates and the lattice size cannot reach the presumed signaling threshold (Fig. 4B) [13, 101]. This isoform-mismatch chain-termination model can explain the recognition initiation process of self and non-self discrimination mediated by the extracellular domains of clustered Pcdhs.
Intracellular Signaling of Clustered Pcdhs Leads to Cytoskeletal Rearrangement and Morphological Remodeling
The intracellular domains of the Pcdhα and Pcdhγ isoforms contain a respective common membrane-distal region encoded by constant exons that is shared by all isoforms from the same cluster [11, 14]. The Pcdhα and Pcdhγ isoforms are cleaved by metalloproteinase and subsequently by γ-secretase to generate a soluble extracellular fragment and an intracellular fragment that may function locally or translocate into the cell nucleus [106–109]. This proteolytic process requires endocytosis and is regulated during animal development and neuronal differentiation [110].
The Pcdhα and Pcdhγ proteins can bind and inhibit two cell-adhesion kinases, FAK (focal adhesion kinase) and Pyk2 (proline-rich tyrosine kinase 2), through the cytoplasmic domain (Fig. 4C) [111]. In the mouse hippocampus and cortex, Pcdhα and Pcdhγ regulate dendritic arborization and spine morphogenesis through inhibiting Pyk2 and FAK activity [112–114]. Knockout or knockdown of Pcdhα in hippocampal neurons results in the phosphorylation and activation of Pyk2 [113]. The activation of Pyk2 inhibits Rac1, leading to defects in dendritic and spine morphogenesis. Consistently, knockdown of Pyk2 or overexpression of Rac1 rescues the phenotype caused by Pcdh α or γ knockdown [113]. Pcdhγ knockout induces extensive neuronal apoptosis in the spinal cord [6], which could be related to aberrantly up-regulated Pyk2 activity. Consistent with this, over expression of Pyk2 also induces apoptosis [111]. Together, these data suggest that diverse extracellular signals acting on different Pcdhα and Pcdhγ isoforms converge into the same intracellular pathways through common downstream effectors of Pyk2 and FAK (Fig. 4C).
The common intracellular domain of Pcdhα isoforms, but not Pcdhγ isoforms, contains a conserved peptide WIRS (WAVE-interacting receptor sequence) motif that interacts with the WAVE (Wiskott-Aldrich syndrome family verprolin homologous protein) regulatory complex (WRC) to modulate cytoplasmic actin assembly (Fig. 1B) [115, 116]. Specifically, Pcdhα isoforms (except for Pcdhαc2) regulate cytoskeletal dynamics during cortical neuron migration and dendrite morphogenesis through the WAVE regulatory complex (Fig. 4C) [116]. Overexpression of Pcdhα isoforms (except for Pcdhαc2) rescues the migration defects caused by Pcdhα knockdown and the rescue is abolished by WIRS mutation. In addition, overexpression of WRC subunits also rescues the migration defects of Pcdhα knockdown [116]. Given that Pcdhα forms cis-heterodimers with Pcdh β or γ on the cell surface (Fig. 4A), the Pcdh β and γ isoforms may also modulate the WAVE complex through interacting with Pcdhα (Fig. 4C). Specifically, Pcdh β and γ proteins, together with Pcdhα, may regulate neuronal morphogenesis and dendrite self-avoidance through WAVE dynamics and cytoskeletal rearrangements (Fig. 4C). In summary, the establishment and maintenance of neuronal connectivity and self-avoidance likely require coordinated collaborations between members of all three Pcdh gene clusters.
Concluding Remarks and Future Perspectives
In the central nervous system, individual neurons stochastically express combinatorial sets of clustered Pcdhs. These Pcdh expression profiles constitute diverse cell-surface identity codes through cis-promiscuous pairing and discriminate self from non-self through strict trans-homophilic interactions. Their tremendous diversity is generated by intriguing 3-D genome architecture, stochastic promoter choice balanced by topological insulators, long-range spatial chromatin contacts between distal enhancers and target promoters, and alternative splicing.
Elucidating the regulatory mechanisms of clustered Pcdhs in different cell types throughout the nervous system will be of great importance in deciphering the molecular basis underlying neural-circuit coding. Several lines of investigation of the Pcdh clusters have provided deep insights into various aspects of gene expression mechanisms, from 1-D genomic organization to 2-D epigenetic regulation and 3-D chromatin architecture. However, many important questions remain unanswered. For example, when are Pcdh isoforms chosen to be expressed in neuronal progenitor cells during brain development? What is the mechanistic basis for the epigenetic memory of clustered Pcdh expression profiles? What are the mechanistic differences between the regulation of expression of alternate and C-type isoforms? How do serotonergic neurons selectively express only the Pcdhαc2 gene in a cell-type-specific manner? Finally, how do clustered Pcdhs collaborate with other families of cell-adhesion proteins to specify synaptic connectivity? Answering these questions about neural coding mechanisms in the brain requires interdisciplinary endeavors in the future.
Acknowledgements
We thank Drs J. Li and J. Shou, and other members of our laboratory for critical reading and discussion. This work was supported by Grants from the National Natural Science Foundation of China (31630039 and 31700666), the Ministry of Science and Technology of China (2017YFA0504203 and 2018YFC1004504), and the Science and Technology Commission of Shanghai Municipality (19JC1412500).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Südhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO. Cell adhesion: old and new questions. Trends Cell Biol. 1999;9:M33–M37. [PubMed] [Google Scholar]
- 4.Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 5.Honig B, Shapiro L. Adhesion protein structure, molecular affinities, and principles of cell-cell recognition. Cell. 2020;181:520–535. doi: 10.1016/j.cell.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanes JR, Zipursky SL. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 8.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 9.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, Li H. Revisiting Dscam diversity: lessons from clustered protocadherins. Cell Mol Life Sci. 2019;76:667–680. doi: 10.1007/s00018-018-2951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 12.Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, et al. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasch J, Goodman KM, Noble AJ, Rapp M, Mannepalli S, Bahna F, et al. Visualization of clustered protocadherin neuronal self-recognition complexes. Nature. 2019;569:280–283. doi: 10.1038/s41586-019-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci U S A. 2000;97:3124–3129. doi: 10.1073/pnas.060027397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying G, Wu S, Hou R, Huang W, Capecchi MR, Wu Q. The protocadherin gene Celsr3 is required for interneuron migration in the mouse forebrain. Mol Cell Biol. 2009;29:3045–3061. doi: 10.1128/MCB.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia Z, Guo Y, Tang Y, Xu Q, Li B, Wu Q. Regulation of the protocadherin Celsr3 gene and its role in globus pallidus development and connectivity. Mol Cell Biol. 2014;34:3895–3910. doi: 10.1128/MCB.00760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Haws P, Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genom Res. 2004;14:79–89. doi: 10.1101/gr.1225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zipursky SL, Grueber WB. The molecular basis of self-avoidance. Annu Rev Neurosci. 2013;36:547–568. doi: 10.1146/annurev-neuro-062111-150414. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama T, Yagi T. Regulation of clustered protocadherin genes in individual neurons. Semin Cell Dev Biol. 2017;69:122–130. doi: 10.1016/j.semcdb.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Lefebvre JL. Neuronal territory formation by the atypical cadherins and clustered protocadherins. Semin Cell Dev Biol. 2017;69:111–121. doi: 10.1016/j.semcdb.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 22.Peek SL, Mah KM, Weiner JA. Regulation of neural circuit formation by protocadherins. Cell Mol Life Sci. 2017;74:4133–4157. doi: 10.1007/s00018-017-2572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstein R, Goodman KM, Maniatis T, Shapiro L, Honig B. Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin Cell Dev Biol. 2017;69:140–150. doi: 10.1016/j.semcdb.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, Maniatis T. Writing, reading, and translating the clustered protocadherin cell surface recognition code for neural circuit assembly. Annu Rev Cell Dev Biol. 2018;34:471–493. doi: 10.1146/annurev-cellbio-100616-060701. [DOI] [PubMed] [Google Scholar]
- 25.Canzio D, Maniatis T. The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Curr Opin Neurobiol. 2019;59:213–220. doi: 10.1016/j.conb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 27.Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci U S A. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trisler GD, Schneider MD, Nirenberg M. A topographic gradient of molecules in retina can be used to identify neuron position. Proc Natl Acad Sci U S A. 1981;78:2145–2149. doi: 10.1073/pnas.78.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelman GM. Cell adhesion molecules. Science. 1983;219:450–457. doi: 10.1126/science.6823544. [DOI] [PubMed] [Google Scholar]
- 31.Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Zeng J, Huang L, Wu D, Liu L, Liu Y, et al. Microarray analysis of gene expression changes in Neuroplastin 65-knockout mice: implications for abnormal cognition and emotional disorders. Neurosci Bull. 2018;34:779–788. doi: 10.1007/s12264-018-0251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 34.Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, et al. Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 36.Chun J. Developmental neurobiology: a genetic Cheshire cat? Curr Biol. 1999;9:R651–R654. doi: 10.1016/s0960-9822(99)80415-1. [DOI] [PubMed] [Google Scholar]
- 37.Mombaerts P. Digging for gold in the human genome. Nat Neurosci. 1999;2:686–687. doi: 10.1038/11142. [DOI] [PubMed] [Google Scholar]
- 38.Serafini T. Finding a partner in a crowd: neuronal diversity and synaptogenesis. Cell. 1999;98:133–136. doi: 10.1016/s0092-8674(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 39.Itzkovitz S, Baruch L, Shapiro E, Segal E. Geometric constraints on neuronal connectivity facilitate a concise synaptic adhesive code. Proc Natl Acad Sci U S A. 2008;105:9278–9283. doi: 10.1073/pnas.0712207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genom Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, et al. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, et al. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Yi Chuan. 2016;38:323–336. doi: 10.16288/j.yczz.16-037. [DOI] [PubMed] [Google Scholar]
- 47.Lu Y, Shou J, Jia Z, Wu Y, Li J, Guo Y, et al. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell. 2019;10:914–920. doi: 10.1007/s13238-019-00656-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia Z, Li J, Ge X, Wu Y, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genom Biol. 2020;21:75. doi: 10.1186/s13059-020-01984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2011;108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 2005;169:2179–2188. doi: 10.1534/genetics.104.037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144:579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM. Gene conversion and the evolution of protocadherin gene cluster diversity. Genom Res. 2004;14:354–366. doi: 10.1101/gr.2133704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, Amemiya CT, et al. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genom Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tada MN, Senzaki K, Tai Y, Morishita H, Tanaka YZ, Murata Y, et al. Genomic organization and transcripts of the zebrafish Protocadherin genes. Gene. 2004;340:197–211. doi: 10.1016/j.gene.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 57.Yu WP, Yew K, Rajasegaran V, Venkatesh B. Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts. BMC Evol Biol. 2007;7:49. doi: 10.1186/1471-2148-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang XJ, Li S, Ravi V, Venkatesh B, Yu WP. Identification and comparative analysis of the protocadherin cluster in a reptile, the green anole lizard. PLoS One. 2009;4:e7614. doi: 10.1371/journal.pone.0007614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Etlioglu HE, Sun W, Huang Z, Chen W, Schmucker D. Characterization of a single genomic locus encoding the clustered protocadherin receptor diversity in Xenopus tropicalis. G3 (Bethesda) 2016;6:2309–2318. doi: 10.1534/g3.116.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodman KM, Rubinstein R, Dan H, Bahna F, Mannepalli S, Ahlsen G, et al. Protocadherin cis-dimer architecture and recognition unit diversity. Proc Natl Acad Sci U S A. 2017;114:E9829–E9837. doi: 10.1073/pnas.1713449114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu WP, Rajasegaran V, Yew K, Loh WL, Tay BH, Amemiya CT, et al. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: a comparative analysis of the protocadherin cluster. Proc Natl Acad Sci U S A. 2008;105:3819–3824. doi: 10.1073/pnas.0800398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524:220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Styfhals R, Seuntjens E, Simakov O, Sanges R, Fiorito G. In silico Identification and expression of protocadherin gene family in octopus vulgaris. Front Physiol. 1905;2018:9. doi: 10.3389/fphys.2018.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet. 2017;49:1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nichols MH, Corces VG. A CTCF code for 3D genome architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Golan-Mashiach M, Grunspan M, Emmanuel R, Gibbs-Bar L, Dikstein R, Shapiro E. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucl Acids Res. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 71.Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen M, Geeven G, et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet. 2018;50:1151–1160. doi: 10.1038/s41588-018-0161-5. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Shou J, Guo Y, Tang Y, Wu Y, Jia Z, et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol. 2015;7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell. 2018;71(498–509):e494. doi: 10.1016/j.molcel.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 74.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toyoda S, Kawaguchi M, Kobayashi T, Tarusawa E, Toyama T, Okano M, et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82:94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Mountoufaris G, Chen WV, Hirabayashi Y, O’Keeffe S, Chevee M, Nwakeze CL, et al. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science. 2017;356:411–414. doi: 10.1126/science.aai8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wada T, Wallerich S, Becskei A. Stochastic gene choice during cellular differentiation. Cell Rep. 2018;24:3503–3512. doi: 10.1016/j.celrep.2018.08.074. [DOI] [PubMed] [Google Scholar]
- 78.Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, et al. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin alpha promoter choice. Cell. 2019;177(639–653):e615. doi: 10.1016/j.cell.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, et al. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res. 2017;27:1365–1377. doi: 10.1038/cr.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen K, Hu J, Moore DL, Liu R, Kessans SA, Breslin K, et al. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc Natl Acad Sci U S A. 2015;112:E3535–E3544. doi: 10.1073/pnas.1504232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan YP, Li S, Jiang XJ, Loh W, Foo YK, Loh CB, et al. Regulation of protocadherin gene expression by multiple neuron-restrictive silencer elements scattered in the gene cluster. Nucl Acids Res. 2010;38:4985–4997. doi: 10.1093/nar/gkq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Justice M, Carico ZM, Stefan HC, Dowen JM. A WIZ/cohesin/CTCF complex anchors DNA loops to define gene expression and cell identity. Cell Rep. 2020;31:107503. doi: 10.1016/j.celrep.2020.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Isbel L, Prokopuk L, Wu H, Daxinger L, Oey H, Spurling A, et al. Wiz binds active promoters and CTCF-binding sites and is required for normal behaviour in the mouse. Elife. 2016;5:e15082. doi: 10.7554/eLife.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A. 2010;107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Almenar-Queralt A, Merkurjev D, Kim HS, Navarro M, Ma Q, Chaves RS, et al. Chromatin establishes an immature version of neuronal protocadherin selection during the naive-to-primed conversion of pluripotent stem cells. Nat Genet. 2019;51:1691–1701. doi: 10.1038/s41588-019-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kallenbach S, Khantane S, Carroll P, Gayet O, Alonso S, Henderson CE, et al. Changes in subcellular distribution of protocadherin gamma proteins accompany maturation of spinal neurons. J Neurosci Res. 2003;72:549–556. doi: 10.1002/jnr.10618. [DOI] [PubMed] [Google Scholar]
- 88.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, et al. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci. 2003;23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, et al. Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci. 2005;29:603–616. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Miralles CP, Taylor MJ, Bear J, Jr, Fekete CD, George S, Li Y, et al. Expression of protocadherin-gammaC4 protein in the rat brain. J Comput Neurol. 2020;528:840–864. doi: 10.1002/cne.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5:e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, Yagi T. Single-neuron diversity generated by Protocadherin-beta cluster in mouse central and peripheral nervous systems. Front Mol Neurosci. 2012;5:90. doi: 10.3389/fnmol.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 94.Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, et al. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. 2006;281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 95.Tan L, Xing D, Daley N, Xie XS. Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat Struct Mol Biol. 2019;26:297–307. doi: 10.1038/s41594-019-0205-2. [DOI] [PubMed] [Google Scholar]
- 96.Chen WV, Nwakeze CL, Denny CA, O’Keeffe S, Rieger MA, Mountoufaris G, et al. Pcdhalphac2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science. 2017;356:406–411. doi: 10.1126/science.aal3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katori S, Noguchi-Katori Y, Okayama A, Kawamura Y, Luo W, Sakimura K, et al. Protocadherin-alphaC2 is required for diffuse projections of serotonergic axons. Sci Rep. 2017;7:15908. doi: 10.1038/s41598-017-16120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murata Y, Hamada S, Morishita H, Mutoh T, Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J Biol Chem. 2004;279:49508–49516. doi: 10.1074/jbc.M408771200. [DOI] [PubMed] [Google Scholar]
- 99.Han MH, Lin C, Meng S, Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol Cell Proteomics. 2010;9:71–83. doi: 10.1074/mcp.M900343-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goodman KM, Rubinstein R, Thu CA, Mannepalli S, Bahna F, Ahlsen G, et al. gamma-Protocadherin structural diversity and functional implications. Elife. 2016;5:e20930. doi: 10.7554/eLife.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, et al. Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell. 2015;163:629–642. doi: 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nicoludis JM, Lau SY, Scharfe CP, Marks DS, Weihofen WA, Gaudet R. Structure and sequence analyses of clustered protocadherins reveal antiparallel interactions that mediate homophilic specificity. Structure. 2015;23:2087–2098. doi: 10.1016/j.str.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsen G, et al. Structural basis of diverse homophilic recognition by clustered alpha- and beta-protocadherins. Neuron. 2016;90:709–723. doi: 10.1016/j.neuron.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicoludis JM, Vogt BE, Green AG, Scharfe CP, Marks DS, Gaudet R. Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4. Elife. 2016;5:e18449. doi: 10.7554/eLife.18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nicoludis JM, Green AG, Walujkar S, May EJ, Sotomayor M, Marks DS, et al. Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis. Proc Natl Acad Sci U S A. 2019;116:17825–17830. doi: 10.1073/pnas.1821063116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haas IG, Frank M, Veron N, Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. J Biol Chem. 2005;280:9313–9319. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- 107.Hambsch B, Grinevich V, Seeburg PH, Schwarz MK. {gamma}-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J Biol Chem. 2005;280:15888–15897. doi: 10.1074/jbc.M414359200. [DOI] [PubMed] [Google Scholar]
- 108.Reiss K, Maretzky T, Haas IG, Schulte M, Ludwig A, Frank M, et al. Regulated ADAM10-dependent ectodomain shedding of gamma-protocadherin C3 modulates cell-cell adhesion. J Biol Chem. 2006;281:21735–21744. doi: 10.1074/jbc.M602663200. [DOI] [PubMed] [Google Scholar]
- 109.Bonn S, Seeburg PH, Schwarz MK. Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Mol Cell Biol. 2007;27:4121–4132. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buchanan SM, Schalm SS, Maniatis T. Proteolytic processing of protocadherin proteins requires endocytosis. Proc Natl Acad Sci U S A. 2010;107:17774–17779. doi: 10.1073/pnas.1013105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen J, Lu Y, Meng S, Han MH, Lin C, Wang X. alpha- and gamma-Protocadherins negatively regulate PYK2. J Biol Chem. 2009;284:2880–2890. doi: 10.1074/jbc.M807417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garrett AM, Schreiner D, Lobas MA, Weiner JA. Gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74:269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Suo L, Lu H, Ying G, Capecchi MR, Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol. 2012;4:362–376. doi: 10.1093/jmcb/mjs034. [DOI] [PubMed] [Google Scholar]
- 114.Keeler AB, Schreiner D, Weiner JA. Protein Kinase C Phosphorylation of a gamma-protocadherin C-terminal lipid binding domain regulates focal adhesion kinase inhibition and dendrite arborization. J Biol Chem. 2015;290:20674–20686. doi: 10.1074/jbc.M115.642306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan L, Lu Y, Shen X, Shao H, Suo L, Wu Q. Alpha protocadherins and Pyk2 kinase regulate cortical neuron migration and cytoskeletal dynamics via Rac1 GTPase and WAVE complex in mice. Elife. 2018;7:e35242. doi: 10.7554/eLife.35242. [DOI] [PMC free article] [PubMed] [Google Scholar]