Figure 2: Origins of PRSS56 c.1066dupC in a mutational hotspot.
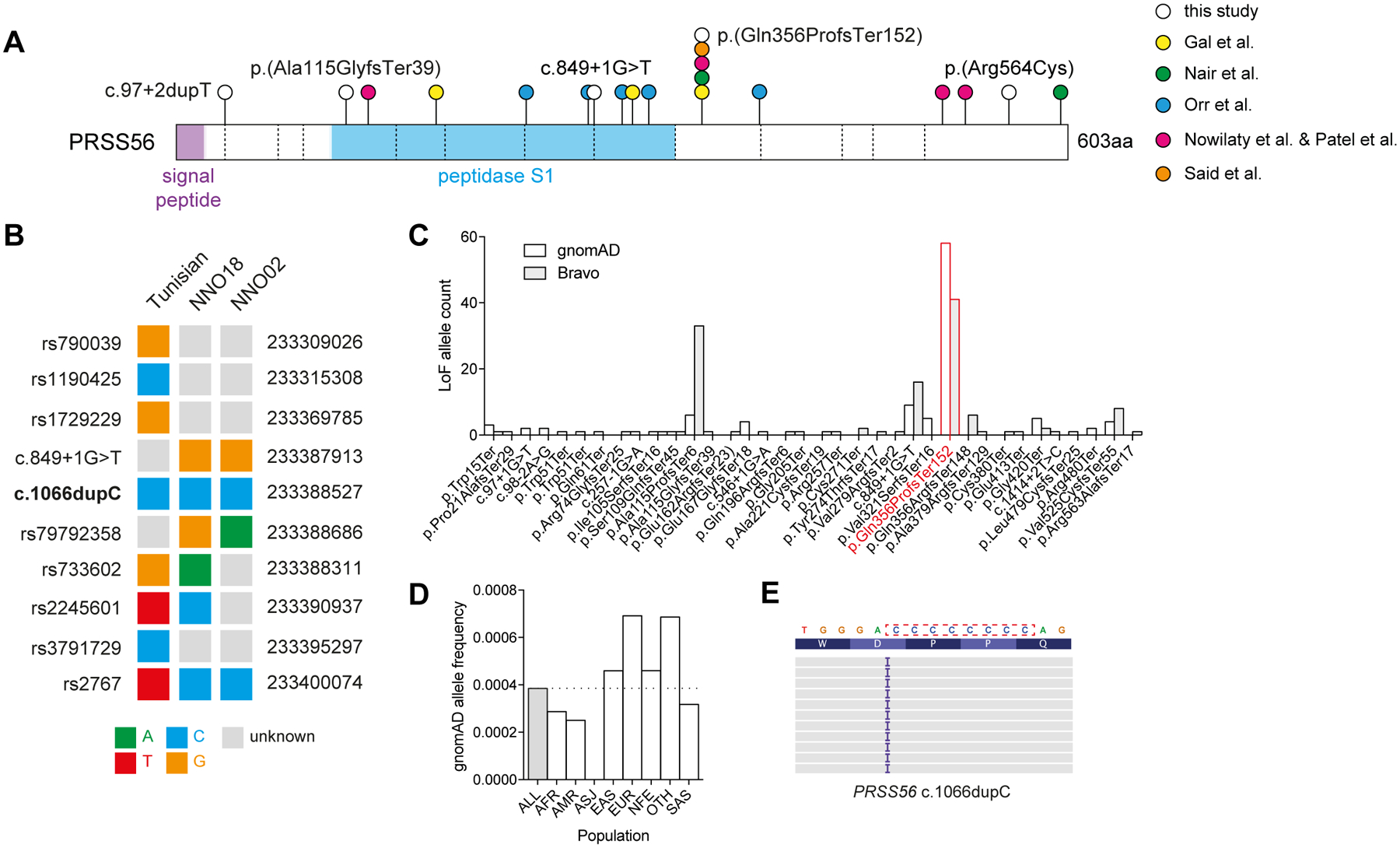
(A) PRSS56 protein schematic with variants identified in this study (white circles), or previous studies (coloured circles). Dashed vertical lines indicate exon boundaries. (B) Haplotype structure surrounding the PRSS56 c.1066dupC variant in a previously reported Tunisian founder, and two unrelated probands. (C) Frequency of PRSS56 loss-of-function variants (nonsense, essential splice, frameshift) reported in gnomAD r2.0.2 and Bravo (TOPMed Freeze5) collections. The c.1066dupC variant is highlighted in red. (D) Frequency of c.1066dupC across multiple ancestries within gnomAD. (E) IGV representation of the left-shifted c.1066dupC variant within a cytosine mononucleotide repeat.
