Abstract
DNA damage repair (DDR) pathways are essential to ensure the accurate transmission of genetic material. However, different endogenous and exogenous factors challenge genomic integrity. Mechanisms involved in the alterations of DDR pathways mainly include genetic inactivation and epigenetic mechanisms. The development and progression of carcinomas are closely associated with DDR pathway aberrations, including the epigenetic silencing of gene O6-alkylguanine-DNA methyltransferase (MGMT); deficiencies of mismatch repair (MMR) genes, including MutL homolog 1 (MLH1), MutS protein homologue (MSH)-2 (MSH2), MSH6, and PMS1 homolog 2; the mismatch repair system component (PMS2); and mutations of homologous recombination repair (HRR) genes, such as the breast cancer susceptibility gene 1/2 (BRCA1/2). Understanding the underlying mechanisms and the correlations between alterations to DDR pathways and cancer could improve the efficacy of antitumor therapies. Emerging evidence suggests that survival is higher in patients with DDR-deficient tumors than in those with DDR-proficient tumors. Thus, DDR alterations play a predictive and prognostic role in anticancer therapies. Theoretical studies on the co-administration of DDR inhibitors and other anticancer therapies, including chemotherapy, radiotherapy, immunotherapy, endocrine therapy, and epigenetic drugs, hold promise for cancer treatments. In this review, we focus on the basic mechanisms, characteristics, current applications, and combination strategies of DDR pathways in the anticancer field.
Keywords: Anticancer therapy, cancer, DNA damage repair pathway (DDR pathway), DNA repair, immunotherapy
Background
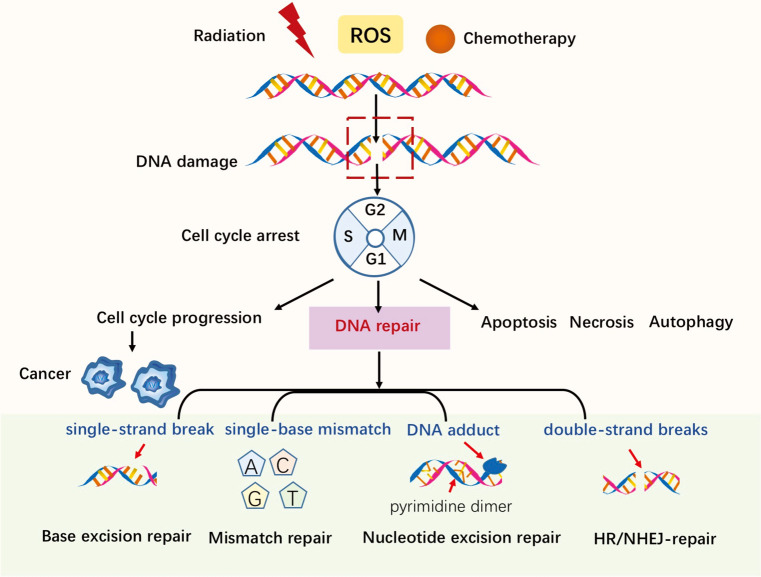
The protection of genomic integrity is necessary if the accurate transmission of genetic information to the next generation is to be ensured. Both endogenous and exogenous factors can challenge genomic integrity; for example, endogenous reactive oxygen species (ROS) induced by normal cellular-metabolism can threaten genomic integrity, while exogenous antitumor therapies, including ionizing radiation and chemotherapies, can cause DNA damage (see Figure 1). Mediated by mechanisms such as post-translational modification and epigenetics, DNA damage repair (DDR) pathways help to maintain genomic stability (1). Alterations of DDR-related genes are closely correlated with cancer cells’ deregulation of DNA repair; thus, an understanding of the mechanisms of how DDR works, along with its characteristics and applications, may be crucial to understanding cancer development and further exploring DDR-related anti-cancer treatments.
Figure 1.
DNA damage and main DNA repair pathways.
DDR pathways
When DNA damage occurs, DNA repair machinery can be recruited; however, severe genotoxic insults in cells may cause apoptosis or necrosis (see Figure 1). Autophagy helps to remove damaged or non-functional organelles (2). Checkpoints are compositions of DNA damage responses. The detection of DNA damage can help to activate checkpoints to ensure faithful DNA reproduction, preventing the progression to the next cell cycle phase. However, cancer is capable of falsely activating checkpoints so that damage continues (3). Here, we briefly examine the main DDR-related pathways and their characteristics. The main components of the DDR system include direct reversal/repair (DR) pathway, base excision repair (BER) pathway, mismatch repair (MMR) pathway, nucleotide excision repair (NER) pathway, non-homologous end joining (NHEJ) pathway, and homologous recombination repair (HRR) pathway (4).
DNA damage can be repaired directly. O6-alkylguanine-DNA methyltransferase (MGMT) was the first DDR-related gene to be studied (5). MGMT can correct DNA damage by removing certain alkyl groups from impaired thymine or guanine bases, without removing the damaged base itself. By binding to an alkyl group, MGMT can inactivate and degrade proteins. Meanwhile, the BER pathway contributes to some prevalent damage as a result of oxidation, deamination, and alkylation. This pathway is crucial to the maintenance of genomic integrality, as a mispairing left unchecked by BER can lead to mutations (6). The MMR pathway corrects mismatches of single-base-pairs (A-G, T-C) and misaligned short nucleotide repeats (7), which can result in frameshift mutations if not repaired and that usually occur during the DNA replication S phase. Repair is accomplished by the MutS protein homologue (MSH)-2: MSH6 or MSH2: MSH3 heterodimer. MutL homolog 1 (MLH1) is recruited as a damage sensor to scaffold protein and to help determine definite strand errors. Other MutL homologues also exist. Similar to MutS, MutL also forms dimers; that is, MutLα [MLH1-PMS1 homolog 2, mismatch repair system component (PMS2)], MutLβ (MLH1-PMS1) and MutLγ (MLH1-MLH3) (8). Notably, MMR deficiency can make malignant cells sensitive to chemotherapeutic drugs; for example, Martin et al. found that methotrexate could induce oxidative DNA damage and was selectively lethal to tumor cells with MSH2 defects (7). The NER pathway is responsible for lesions caused by chemical agents or ultraviolet radiation (9). The main mechanisms of double-strand break (DSB) repair include NHEJ and HRR (in which the HRR pathway is regarded as the best guarantee of genomic stability) (10). HRR is the best guarantee of genomic stability; however, it occurs only during the S and G2 phases of the cell cycle, as it depends on the sister chromatid as a template for repair. Thus, NHEJ is the main DSB repair pathway in eukaryotes, even though it is an error-prone pathway (10).
Basic mechanisms of DDR pathway deficiency in cancer
The mechanisms behind DDR defect in cancer mainly include genetic inactivation and epigenetics (11,12). Genetic inactivation is the most typical method of DDR deficiency and can alter DNA sequences through germline or somatic mutations. For example, hereditary nonpolyposis colorectal cancer Lynch syndrome is caused by germline inactivation. Some patients with uncommon hereditary cancer syndromes (such as ataxia-telangiectasia) also show biallelic inactivation (13,14). A pan-cancer analysis by Robinson’s group reported that 12% of patients had pathogenic germline mutations, and 75% of these germline variants were DDR-related mutations (15). Somatic mutations can also mediate defects of many DDR-related genes (16).
Multiple epigenetic mechanisms are involved in the regulation of DDR pathways, such as DNA methylation, nucleosome remodeling, and histone modification. In the DR pathway, MGMT promoter methylation can lead to the loss of somatic function (17). Epigenetic silencing is important in genomic instability. The microsatellite instability (MSI) phenotype is closely correlated with the epigenetic silencing of MLH1. In a colonic cancer related study, Nakagawa et al. observed MLH1 hypermethylation before the occurrence of MSI (18). The MGMT promoter methylation also illustrates the importance of epigenetic silencing. The epigenetic silencing of MGMT represents a strong predictive and prognostic biomarker for alkylating agents in colorectal cancer and glioblastoma (19). Additionally, DDR is linked to histone post-translational modifications, such as acetylation, phosphorylation, methylation, and ubiquitylation. Isocitrate dehydrogenase mutations can induce HRR deficiency by inhibiting the α-ketoglutarate dependent dioxygenases that are related to epigenetic reprogramming in cells (20). Additionally, several viruses have been found to play essential roles in the regulation of DDR pathways (21).
Features of DDR pathway alterations in cancer
Alterations to the DR pathway
DDR alterations are closely correlated with various cancers (see Table 1). Epigenetic silencing is the most prevalent alteration to the DR pathway. MGMT inactivation by promoter hypermethylation has been detected in some cancer types, such as colorectal carcinomas, gliomas, non-small cell lung carcinomas, head and neck carcinomas, and lymphomas (37). A study based on The Cancer Genome Atlas (TCGA) pan-cancer data analyzed DDR alterations across 33 types of cancer. The results revealed the epigenetic silencing of direct genes MGMT, exonuclease 5 and alpha-ketoglutarate–dependent dioxygenase alkB homolog 3 (ALKBH3) in approximately 20% of the samples (38). The epigenetic silencing of genes MGMT and ALKBH3 constitutes DNA repair in multiple cancer types; however, some types of cancer are vulnerable to DNA damage (39,40). Additionally, in gastrointestinal, lymphoid, and central nervous cancers, alterations to DR-related genes are all related to a high tumor mutation burden (TMB) (38). Thus, these alterations may be used to identify a subset of patients who respond to immunotherapies.
Table 1. DDR pathway alterations in cancers.
| Reference | DDR pathway | Genomic alteration | Tumor type | Size of assessable cases | Prevalence |
|---|---|---|---|---|---|
| Hegi et al. (22) | DR | MGMT promoter methylation | Glioblastoma | 206 | 45% |
| Ruemmele et al. (23) | MMR | Loss of MLH1, MSH2 and MSH6 | Ampullary carcinoma | 144 | 10% |
| Hampel et al. (24) | MMR | Mutations of MLH1, MSH2, MSH6, and PMS2 | Colorectal cancer | 1,066 | 13% |
| Hampel et al. (25) | MMR | Mutations of MLH1, MSH2, MSH6 | Endometrial cancer | 543 | 22% |
| TCGA, Kandoth et al. (26) | MMR | MLH1 promoter methylation | Endometrial cancer | 373 | MSI-H: 30% |
| TCGA (27) | MMR | hypermethylation at the MLH1 promoter | Gastric cancer | 295 | 22% |
| Goumard et al. (28) | MMR | Mutations of MSH2 or MLH1 | Hepatocellular carcinoma | 122 | MSI-H: 0%; MSI: 26.2% |
| Alsop et al. (29) | HRR | BRCA1/2 deficiency | Ovarian carcinoma | 1,001 | 14% |
| Walsh et al. (30) | HRR | BRCA1/2 deficiency | Ovarian carcinomas | 360 | 18% |
| Rennert eet al. (31) | HRR | BRCA1/2 deficiency | Breast cancer | 1,545 | 10% |
| TCGA, Kandoth et al. (26) | BER | POLE mutation | Endometrial cancer | 373 | 7% |
| TCGA (32) | BER | POLE mutation | Colorectal carcinoma | 224 | 5.8% |
| Scarpitta et al. (33) | BER | MUTYH mutation | Male breast cancer | 81 | 1.2% |
| Stoffel et al. (34) | BER | MUTYH mutation | Colorectal cancer | 430 | 1.86% |
| Yap et al. (35) | NER | ERCC2 mutation | Muscle-invasive bladder cancer | 43 | 7% |
| TCGA (36) | NER | ERCC2 mutation | Urothelial bladder carcinoma | 131 | 12% |
The column entitled “Size of assessable cases” refers to the number of all assessable cases enrolled in each study, including cases with or without the genomic alteration in the DDR pathway. The column entitled “Prevalence” refers to the proportion of the cases of the genomic alteration observed in all assessable cases. DDR, DNA damage repair; DR, direct reversal/repair; MMR, mismatch repair; HRR, homologous recombination repair; BER, base excision repair; NER, nucleotide excision repair.
BER pathway defects
The BER pathway repairs multiple types of endogenous DNA damage, and many important players are involved in this pathway. Apurinic/apyrimidinic endonuclease 1, an important component protein, is a central player in BER, and is responsible for more than 95% of this process (41). The BER mutation rate is low in the general population. Indeed, only a very small number of patients have been found to have mutY homolog (MUTYH) biallelic mutations, which involve the encoding of DNA glycosylase in the BER pathway. Prominent immune cell infiltration and increased TMB are associated with these kinds of tumors (42,43). However, very few clinical reports have discussed these patients’ sensitivity to programmed death 1 (PD-1) blockage.
MMR deficiency
MMR deficiency is a potential biomarker for immune checkpoint inhibitor (ICI)-based immunotherapy. The germline sequencing of MMR genes (MLH1, MSH2, MSH6, and PMS2) is commonly performed to detect MMR deficiency in clinical practice (44). High tumor burden has been found to be a feature of MMR-deficient cancers. The MSI phenotype is formed as a consequence of large numbers of mutations accumulating in MMR-defective tumors, secondary to short repeats of DNA sequences and the substitution of a single nucleotide. A 100- to 1,000-fold increase in the mutation rates of frameshift and missense mutations has been reported in MSI-high cancers (45).
Individuals with lynch syndrome are prone to develop colorectal and other cancers due to the germline mutations of MMR genes, or the loss of expressed proteins by epigenetic alterations (46). MMR defects have also been observed in cancers such as gastrointestinal and endometrial carcinomas (47,48). In addition to high TMB, this tumor phenotype is also associated with prominent lymphocyte, and has an elevated expression of PD-1 and programmed death ligand 1 (PD-L1) (49). Furthermore, patients with MMR-defective tumors may have higher responses to ICIs. Le et al. conducted a study of 12 different cancer types, and found that the PD-1 blockade had durable efficacy in patients with MMR-defective tumors. Objective responses in radiography were observed for 53% of patients in this study, and complete responses were observed in 21% of patients (44).
NER pathway aberrations
Somatic alterations in the NER pathway, especially mutations of excision repair cross complement (ERCC) genes, are common in cancers. In a study of non-small cell lung cancer (NSCLC), Aiello et al. found that patients with ERCC1 alteration had a high response to nivolumab (50). Xeroderma pigmentosum (XP) genes (XP-A to -G and variants), which are associated with XP, a genetic syndrome characterized by extreme sun sensitivity that leads to a higher risk of skin cancer, are also important in the NER pathway (51). This syndrome is extremely rare in clinic; however, durable and dramatic response to ICIs has been observed in non-melanomatous and melanomatous XP-related skin cancers (52-54). Thus, NER pathway aberrations might predict the prognosis of cancer patients treated with immunotherapy.
NHEJ pathway aberrations
A number of defects of the NHEJ pathway have been shown to be associated with cancers. In the NHEJ pathway, the main proteins to mediate DSB repair include X-ray repair cross-complementing protein 5 (XRCC5) (Ku80), XRCC6 (Ku70), XRCC4, DNA ligase 4 (LIG4), and the XRCC four-like factor (XLF) (55). In NHEJ, the epigenetic silencing of XRCC5 has been identified as being associated with lung cancer, and the epigenetic inactivation of XRCC6 has been found to be related to colorectal, breast, and lung cancer (56,57). Additionally, some aberrations of XRCC4 and LIG4 can promote genomic instability and increase the radiosensitivity of tumor cells. By studying genetic polymorphism, Gomes et al. discovered that some variants of NHEJ genes might contribute to the susceptibility of thyroid cancer (58). The NHEJ polymorphic variants (in particular LIG4 rs1805388) are capable of modulating the risk of radiation pneumonitis in NSCLC patients treated with radiotherapy (55).
HRR deficiency
The relationship between HRR and the DDR system has been well studied. This type of DNA repair is aptly named, as in a somewhat homologous manner, it usually uses the sister chromatid as a template for reproducing lost or damaged bases. The role of HRR mutations has long been recognized as being related to tumor susceptibility. For example, mutations of the breast cancer susceptibility gene 1/2 (BRCA1/2) in the HRR pathway are associated with hereditary breast and ovarian cancer syndromes. Additionally, alterations of the partner and localizer of BRCA2 (PALB2) can also be observed in cancers (59). Somatic and germline mutations of HRR-related genes have been detected in extensive tumor types (60). Defective HRR pathways are able to accelerate aging, ultimately increasing the risk of cancer. Interestingly, research has shown that HRR deficiency plays an important role in predicting patients’ response to immunotherapy. A melanoma study by Hugo et al. indicated that responding tumors were enriched by BRCA2 mutation during the treatment of the PD-1 inhibitor (61).
Potential of DDR pathway alterations as biomarkers in anticancer therapies
Genomic and proteomic TCGA analyses have identified the frequency of different gene mutations in DDR pathways in different types of cancers. Kandoth et al. identified the frequencies of MSI in DNA polymerase epsilon (POLE) mutations in endometrial and colorectal cancers to be 40% and 11%, respectively, and the mutation rates to be 7% and 3%, respectively (26). Alsop et al. found that germline mutations of BRCA1/2 improved survival in a cohort of 1001 patients with ovarian cancer, and had a mutation rate of 14.1% (29). Hegi et al. conducted an early clinical randomized trial with glioblastoma patients receiving radiotherapy only and patients receiving a combination of radiotherapy and temozolomide, and explored the association between the epigenetic silencing of MGMT and patients’ survival. The results showed that 45% of 206 evaluable patients had methylation of the MGMT promoter. Additionally, glioblastoma patients with the methylated MGMT promoter were found to have longer survival than those without methylated MGMT promoter (22). Thus, the MGMT promoter methylation may be a favorable prognosis factor for predicting survival of glioblastoma patients receiving alkylating agents, and thus should be the subject of further research. Collectively, these findings support the notion that DDR alterations are common in patients with malignancies.
Emerging evidence suggests that patients with DDR mutations may enjoy better clinical treatment efficacy than those with DDR-proficient tumors. After conducting exome and gene sequencing in patients with bladder cancer, Yap et al. found that somatic mutations of one or more DDR-related genes, including the ataxia-telangiectasia mutated (ATM) gene, ERCC2, BRCA1, BRCA2, Fanconi anemia group D2, and PALB2, were correlated with high somatic mutational loads. Further, they identified a relationship between DDR alterations and clinical recurrence-free survival (RFS), and conducted a Kapan-Meier analysis that showed that carrying DDR-deficient mutations enhanced patients’ RFS (35). Ruemmele et al. conducted a multicenter clinical trial with 120 patients with ampullary adenomas and 170 patients with adenocarcinomas. Following an immunohistochemical analysis, patients were divided into one of the following three groups: (I) MSI-high; (II) MSI-low; and (III) the microsatellite-stable group. The results showed that patients in the MSI-high carcinoma group had higher overall survival rates that patients in the MSI-low carcinoma and microsatellite-stable groups (23). These alterations can produce novel tumor-related antigens that can increase the immune system’s targeting of tumor cells.
When delayed or faulty repair occurs, genetic and tumor microenvironment alterations can appear. Modern immunotherapy that blocks tumor-immunocyte connections can cause effective response in many cancers (62). The beginning of the ICI therapy era was marked by clinical trials revealing that a cytotoxic T lymphocyte antigen 4 blocker, ipilumimab, could confer survival benefits to metastatic melanoma patients (63,64). Numerous studies have shown that DDR considerably effects tumor sensitivity and patients’ response to ICIs. Tumor neo-antigens arise from tumor-specific and non-synonymous somatic DNA mutations that can enhance tumor immunogenicity in the immune system (65). Thus, high mutational loads and predicted tumor antigens play important roles in patients’ responses to ICI-based immunotherapy, with various clinical settings having replicated these findings (66-68). Additionally, high TMB has recently been recognized as a potential biomarker in predicting the efficacy of ICI-based immunotherapy in lung cancer (69,70).
MMR defects and MSI-high phenotypes are associated with high degrees of genomic instability (71). In metastatic colorectal cancer, the co-administration of nivolumab and ipilimumab has been shown to have durable clinical benefits for patients with MSI-high tumors (72). Accumulating evidence shows that cancers with HRR deficiency have elevated immunogenicity. In relation to high-grade serous ovarian cancers, patients with BRCA1/2 mutations have been found to have higher neoantigen load than HRR-proficient patients. Additionally, increased PD-1/PD-L1 expression and tumor-infiltrating lymphocytes (TILs) have been observed in patients with BRCA1/2 mutations (73). TIL accumulation has also been observed in breast cancer patients with DDR deficiency (74,75). In wild-type BRCA1/2 ovarian cancer, tumors with genomic alterations, including phosphatase and tensin homologue deleted on chromosome 10 (PTEN) deletion, BRCA1 promoter hypermethylation, ATM. and ATM and Rad3-related (ATR) mutations, have been associated with higher predictive neo-antigen levels than HRR-proficient tumors (76). Thus, DDR pathway alterations may lead to higher mutational loads and higher lymphocyte infiltration, and increase patients’ response to immunotherapy.
Combination strategies
Combination of DDR inhibitors with DNA-damaging agents
The combination of DDR inhibitors and chemotherapies is common in clinical practice (see Table 2). Nevertheless, in many cases, this combination is challenged by the presence of toxicity, such as myelosuppression caused by olaparib–carboplatin combination (77). However, in a phase 2 trial of advanced gastric cancer patients with ATM loss, olaparib showed promising results when combined with chemotherapy (paclitaxel) (78), but, based on a AstraZeneca press release on May 18, 2016, the survival benefit did not significantly improve in a phase 3 GOLD trial. Careful consideration needs to be given to the dose used and the sequence of the administration.
Table 2. Clinical combination therapies of DDR inhibitors with other anticancer therapies.
| Intervention | Cancer type | Phase | Clinical trial ID |
|---|---|---|---|
| DDR inhibitor + chemotherapy | |||
| Abt-888 + cyclophosphamide | Ovarian, breast, and fallopian tube cancer | Phase 2 | NCT01306032 |
| Olaparib + carboplatin | Breast, ovarian, uterine, and cervical cancer | Phase 1 | NCT01237067 |
| Iniparib + paclitaxel | Breast cancer | Phase 2 | NCT01204125 |
| Rucaparib + cisplatin | Breast cancer | Phase 2 | NCT01074970 |
| DDR inhibitor + immunotherapy | |||
| Rucaparib +atezolizumab | Solid tumor | Phase 2 | NCT04276376 |
| Olaparib + durvalumab | Endometrial cancer | Phase 2 | NCT03951415 |
| Olaparib + pembrolizumab | Prostatic neoplasms | Phase 3 | NCT03834519 |
| Niraparib + TSR-042 | Endometrial cancer | Phase 2 | NCT03016338 |
| Talazoparib + avelumab | Ovarian cancer | Phase 3 | NCT03642132 |
| Rucaparib + nivolumab | Biliary tract cancer | Phase 2 | NCT03639935 |
| DDR inhibitor + radiotherapy | |||
| Veliparib + radiation therapy + temozolomide | Malignant glioma | Phase 2 | NCT03581292 |
| Olaparib + radiotherapy | Breast cancer | Phase 1 | NCT02227082 |
| Olaparib + radiotherapy | Head and neck neoplasms | Phase 1 | NCT02229656 |
| Olaparib + radiation therapy | Triple negative breast cancer | Phase 1 | NCT03109080 |
| DDR inhibitor + endocrine therapy | |||
| Niraparib + enzalutamide | Metastatic prostate cancer | Phase 1 | NCT02500901 |
| Olaparib + degarelix | Prostate cancer | Phase 1 | NCT02324998 |
| Olaparib + abiraterone | Metastatic prostate cancer | Phase 2 | NCT01972217 |
| Veliparib + abiraterone acetate | Prostate cancer | Phase 2 | NCT01576172 |
DDR, DNA damage repair.
Combination of DDR inhibitors with immunotherapy
Poly ADP-ribose polymerase inhibitors (PARPis) are paradigmatic examples of DDR-targeted therapy that can increase DNA damage and stimulate the immune system to recognize cancer cells (79). PARPis can also upregulate PD-L1 expression (80). Furthermore, DNA damage has been studied in relation to the activation of the stimulator of interferon genes (STING) pathway, which can enhance cellular innate immunity and increase the production of type I interferons (IFNs) to activate the antitumor immune response (32). Thus, the combination of PARPis with ICIs represents a promising strategy. At last, some clinical trials have been developed to investigate the efficacy and safety of DDR inhibitor/ICI therapy (see Table 2).
Combination of DDR inhibitors with radiotherapy
Radiation-related DNA damage, such as single-strand breaks and DSBs can cause DDR. This stress can be stressed-mediated by central kinases, including DNA-dependent protein kinase, ATM, and ATR, to facilitate DNA repair. As our understanding of DDR pathways have increased, some radiosensitization methods have been developed, such as NHEJ inhibition by suppressing DNA-PK, or the depletion of ATR, RAD51, and CHK1 by the inhibition of chaperone HSP90 (81-83). Micronuclei can be upregulated when combining DDR inhibitors and radiotherapy. Further, DDR inhibitors can contribute to radiotherapy-related inflammatory response, such as in the radiation-related type I IFN responses induced by ATR inhibitors (84).
Combination of DDR Inhibitors with Endocrine Therapy
The crosstalk between DDR and endocrine-related signaling has gradually grown as an area of more intense research interest. Steroid hormone can transcriptionally regulate NHEJ components, thereby promoting NHEJ, and can have both positive and negative effects on HRR in different tumor types (85). PARP1 is important, as it supports the transcriptional function of androgen, which is critical for the transcriptional activation of oncogenic TMPRSS2-ERG protein in approximately 50% of prostate cancer cases (86). In addition, combining the blockage of PARP and androgen-receptor signaling has been shown to better delay tumor progression compared to monotherapy in xenograft mice with prostate cancer (87). Thus, future studies should seek to examine the effects of combining PARPi and endocrine therapy in clinical trials.
Combination of DDR inhibitors with epigenetic drugs
Given its special role in epigenetics, DDR could be used as potential strategy to combine DDR inhibitors and epigenetic drugs. Recently, emerging in vivo and in vitro trials have demonstrated that DNA methyltransferase inhibitors (DNMTis) act synergistically with PARPis to induce tumor cell death (88,89). Cytosine analogues 5-azacytidine and 5-aza-2’-deoxycytidine (decitabine), the most used DNMTis, can be incorporated into DNA to prompt the formation of DNMT-DNA adducts, which can lead to DNA demethylation via DNMT1 inhibition (90). Additionally, DNMTis are capable of enhancing PARP1 trapping at the sites of DNA damage, increasing the cytotoxicity of PARP inhibition (88). DNMTis can also induce ROS accumulation and promote PARP activation, which enhances cancer cells’ sensitivity to PARPis (89).
Conclusions
DDR pathways play important roles in different types of cancer. When endogenous or exogenous damage occurs, genomic integrity is challenged. Genetic instability is a specific feature of malignant cells, and alterations to DDR pathways may lead to tumorigenesis. Interestingly, increasing evidence suggests that cancers with DDR mutations may have high mutational loads and neo-antigens that can trigger immune system to fight tumors. Notably, survival rates are higher in patients with DDR-deficient tumors than in those with DDR-proficient tumors. The combination of DDR inhibitors and other anticancer therapies, such as immunotherapy, radiotherapy, and epigenetic agents, appears promising. Alterations to DDR pathways are important in the development of malignancies and may provide significant strategies for antitumor therapies.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was funded in part by a grants from the National Natural Science Foundation of China (No. 81802255), the Shanghai Pujiang Program (No. 17PJD036), the Shanghai Municipal Commission of Health and Family Planning Program (No. 20174Y0131), the National Key Research and Development Project (No. 2016YFC0902300), the Major Disease Clinical Skills Enhancement Program of the Three-Year Action Plan for Promoting Clinical Skills and Clinical Innovation in Municipal Hospitals, the Shanghai Shen Kang Hospital Development Center Clinical Research Plan of SHDC (No. 16CR1001A), the “Dream Tutor” Outstanding Young Talents Program (No. fkyq1901), the Key Disciplines of Shanghai Pulmonary Hospital (No. 2017ZZ02012), and the Shanghai Science and Technology Commission (No. 16JC1405900).
Ethical Statement: The authors are accountable for all aspects of the work and have ensured that questions related to the accuracy or integrity of all aspects of the work have been appropriately investigated and resolved.
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2920
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2920). The authors have no conflicts of interest to declare.
(English Language Editor: L. Huleatt; Quality Control Editor: J. Gray)
References
- 1.Chatterjee N, Walker GC. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ Mol Mutagen 2017;58:235-63. 10.1002/em.22087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo QQ, Wang SS, Zhang SS, et al. ATM-CHK2-Beclin 1 axis promotes autophagy to maintain ROS homeostasis under oxidative stress. EMBO J 2020:e103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Węsierska-Gądek J, Maurer M, Zulehner N, et al. Whether to Target Single or Multiple CDKs for Therapy? That is the Question. J Cell Physiol 2011;226:341-9. 10.1002/jcp.22426 [DOI] [PubMed] [Google Scholar]
- 4.Scarbrough PM, Weber RP, Iversen ES, et al. A Cross-Cancer Genetic Association Analysis of the DNA Repair and DNA Damage Signaling Pathways for Lung, Ovary, Prostate, Breast, and Colo-rectal Cancer. Cancer Epidemiol Biomarkers Prev 2016;25:193-200. 10.1158/1055-9965.EPI-15-0649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerson SL. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer 2004;4:296-307. 10.1038/nrc1319 [DOI] [PubMed] [Google Scholar]
- 6.Çağlayan M. The ligation of pol beta mismatch insertion products governs the formation of promutagenic base excision DNA repair intermediates. Nucleic Acids Res 2020;48:3708-21. 10.1093/nar/gkaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin SA, McCarthy A, Barber LJ, et al. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. Embo Mol Med 2009;1:323-37. 10.1002/emmm.200900040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roesner LM, Mielke C, Fahnrich S, et al. Stable expression of MutLgamma in human cells reveals no specific response to mismatched DNA, but distinct recruitment to damage sites. J Cell Biochem 2013;114:2405-14. 10.1002/jcb.24591 [DOI] [PubMed] [Google Scholar]
- 9.Bret C, Klein B, Moreaux J. Nucleotide excision DNA repair pathway as a therapeutic tar-get in patients with high-risk diffuse large B cell lymphoma. Cell Cycle 2013;12:1811-2. 10.4161/cc.25115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010;79:181-211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JH. Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair. Int J Mol Sci 2019;20:4093. 10.3390/ijms20174093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilié PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA dam-age response in cancer. Nat Rev Clin Oncol 2019;16:81-104. 10.1038/s41571-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlo MI, Ravichandran V, Srinavasan P, et al. Cancer Susceptibility Mutations in Patients With Urothelial Malignancies. J Clin Oncol 2020;38:406-14. 10.1200/JCO.19.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genetics 2006;38:873-5. 10.1038/ng1837 [DOI] [PubMed] [Google Scholar]
- 15.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature 2017;548:297. 10.1038/nature23306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polak P, Kim J, Braunstein LZ, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017;49:1476-86. 10.1038/ng.3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousuf A, Bhat MY, Pandith AA, et al. MGMT gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell Oncol (Dordr) 2014;37:245-52. 10.1007/s13402-014-0179-3 [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa H, Nuovo GJ, Zervos EE, et al. Age-related hypermethylation of the 5' region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res 2001;61:6991-5. [PubMed] [Google Scholar]
- 19.Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol 2015;26:1994-9. 10.1093/annonc/mdv272 [DOI] [PubMed] [Google Scholar]
- 20.Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017;9:eaal2463. 10.1126/scitranslmed.aal2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weitzman MD, Weitzman JB. What's the Damage? The Impact of Pathogens on Path-ways that Maintain Host Genome Integrity. Cell Host Microbe 2014;15:283-94. 10.1016/j.chom.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 23.Ruemmele P, Dietmaier W, Terracciano L, et al. Histopathologic Features and Microsatellite Instability of Cancers of the Papilla of Vater and Their Precursor Lesions. Am J Surg Pathol 2009;33:691-704. 10.1097/PAS.0b013e3181983ef7 [DOI] [PubMed] [Google Scholar]
- 24.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851-60. 10.1056/NEJMoa043146 [DOI] [PubMed] [Google Scholar]
- 25.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary non-polyposis colorectal cancer) among endometrial cancer patients. Cancer Res 2006;66:7810-7. 10.1158/0008-5472.CAN-06-1114 [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network , Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goumard C, Desbois-Mouthon C, Wendum D, et al. Low Levels of Microsatellite Instability at Simple Repeated Sequences Commonly Occur in Human Hepatocellular Carcinoma. Cancer Genomics Proteomics 2017;14:329-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsop K, Fereday S, Meldrum C, et al. BRCA Mutation Frequency and Patterns of Treatment Response in BRCA Mutation-Positive Women With Ovarian Cancer: A Report From the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30:2654-63. 10.1200/JCO.2011.39.8545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:18032-7. 10.1073/pnas.1115052108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rennert G, Bisland-Naggan S, Barnett-Griness O, et al. Clinical outcomes of breast can-cer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med 2007;357:115-23. 10.1056/NEJMoa070608 [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarpitta R, Zanna I, Aretini P, et al. Germline Investigation in DNA Repair Genes of Male Breast Cancer by Next-Generation Sequencing. Breast Cancer Res Treat 2019;178:557-64. 10.1007/s10549-019-05429-z [DOI] [PubMed] [Google Scholar]
- 34.Stoffel EM, Koeppe E, Everett J, et al. Germline Genetic Features of Young Individuals With Colorectal Cancer. Gastroenterology 2018;154:897. 10.1053/j.gastro.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yap KL, Kiyotani K, Tamura K, et al. Whole-Exome Sequencing of Muscle-Invasive Bladder Cancer Identifies Recurrent Mutations of UNC5C and Prognostic Importance of DNA Repair Gene Mutations on Survival. Clin Cancer Res 2014;20:6605-17. 10.1158/1078-0432.CCR-14-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein JN, Akbani R, Broom BM, et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793-7. [PubMed] [Google Scholar]
- 38.Knijnenburg TA, Wang LH, Zimmermann MT, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep 2018;23:239. 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends Biochem Sci 2017;42:206-18. 10.1016/j.tibs.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, Wu J, Ma S, et al. Oncometabolite D-2-Hydroxyglutarate Inhibits ALKBH DNA Repair Enzymes and Sensitizes IDH Mutant Cells to Alkylating Agents. Cell Rep 2015;13:2353-61. 10.1016/j.celrep.2015.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Yang X, Lu X, et al. APE1 deficiency promotes cellular senescence and premature aging features. Nucleic Acids Res 2018;46:5664-77. 10.1093/nar/gky326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen M, de Miranda NFCC, van Puijenbroek M, et al. Colorectal carcinomas in MUTYH-associated polyposis display histopathological similarities to microsatellite unstable carcinomas. BMC Cancer 2009;9:184. 10.1186/1471-2407-9-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viel A, Bruselles A, Meccia E, et al. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. Ebiomedicine 2017;20:39-49. 10.1016/j.ebiom.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther 2018;189:45-62. 10.1016/j.pharmthera.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 46.Hesson LB, Packham D, Kwok CT, et al. Lynch syndrome associated with two MLH1 promoter variants and allelic imbalance of MLH1 expression. Hum Mutat 2015;36:622-30. 10.1002/humu.22785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz-Bañobre J, Goel A. DNA Mismatch Repair Deficiency and Immune Checkpoint Inhibitors in Gastrointestinal Cancers. Gastroenterology 2019;156:890-903. 10.1053/j.gastro.2018.11.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stelloo E, Jansen AML, Osse EM, et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol 2017;28:96-102. 10.1093/annonc/mdw542 [DOI] [PubMed] [Google Scholar]
- 49.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aiello MM, Vigneri PG, Bruzzi P, et al. Excision repair cross complementation group 1 (ERCC-1) gene polymorphisms and response to nivolumab in advanced non-small cell lung cancer (NSCLC). J Clin Oncol 2017;35:3032. 10.1200/JCO.2017.35.15_suppl.3032 [DOI] [Google Scholar]
- 51.Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet 2001;43:71-102. 10.1016/S0065-2660(01)43004-5 [DOI] [PubMed] [Google Scholar]
- 52.Hauschild A, Eichstaedt J, Mobus L, et al. Regression of melanoma metastases and multiple non-melanoma skin cancers in xeroderma pigmentosum by the PD1-antibody pembrolizumab. Eur J Cancer 2017;77:84-7. 10.1016/j.ejca.2017.02.026 [DOI] [PubMed] [Google Scholar]
- 53.Kraemer KH, Tamura D, Khan SG. Pembrolizumab treatment of a patient with xeroderma pigmentosum with disseminated melanoma and multiple nonmelanoma skin cancers. Br J Dermatol 2018;178:1009. 10.1111/bjd.16525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salomon G, Maza A, Boulinguez S, et al. Efficacy of anti-programmed cell death-1 immunotherapy for skin carcinomas and melanoma metastases in a patient with xeroderma pigmentosum. Br J Dermatol 2018;178:1199-203. 10.1111/bjd.16270 [DOI] [PubMed] [Google Scholar]
- 55.Yin M, Liao ZX, Liu ZS, et al. Genetic variants of the nonhomologous end joining gene LIG4 and severe radiation pneumonitis in non small cell lung cancer patients treated with definitive radiotherapy. Cancer 2012;118:528-35. 10.1002/cncr.26214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MN, Tseng RC, Hsu HS, et al. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res 2007;13:832-8. 10.1158/1078-0432.CCR-05-2694 [DOI] [PubMed] [Google Scholar]
- 57.Willems P, Claes K, Baeyens A, et al. Polymorphisms in nonhomologous end-joining genes associated with breast cancer risk and chromosomal radiosensitivity. Genes Chromosomes Cancer 2008;47:137-48. 10.1002/gcc.20515 [DOI] [PubMed] [Google Scholar]
- 58.Gomes BC, Silva SN, Azevedo AP, et al. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol Rep 2010;24:1079-85. [PubMed] [Google Scholar]
- 59.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110-20. 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- 60.Riaz N, Blecua P, Lim RS, et al. Pancancer analysis of bi-allelic alterations in homologous re-combination DNA repair genes. Nat Commun 2017;8:857. 10.1038/s41467-017-00921-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to An-ti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165:35-44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell 2015;27:450-61. 10.1016/j.ccell.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hodi FS, O'Day SJ, McDermott DF, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med 2010;363:711-23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. N Engl J Med 2011;364:2517-26. 10.1056/NEJMoa1104621 [DOI] [PubMed] [Google Scholar]
- 65.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69-74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 66.Gubin MM, Zhang XL, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515:577. 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853. 10.1016/j.ccell.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Łuksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017;551:517. 10.1038/nature24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Overman MJ, Lonardi S, Wong KYM, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colo-rectal Cancer. J Clin Oncol 2018;36:773. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 73.Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016;7:13587-98. 10.18632/oncotarget.7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green AR, Aleskandarany MA, Ali R, et al. Clinical Impact of Tumor DNA Repair Expression and T-cell Infiltration in Breast Cancers. Cancer Immunol Res 2017;5:292-9. 10.1158/2326-6066.CIR-16-0195 [DOI] [PubMed] [Google Scholar]
- 75.Nolan E, Savas P, Policheni AN, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med 2017;9:eaal4922. 10.1126/scitranslmed.aal4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McAlpine JN, Porter H, Kobel M, et al. BRCA1 and BRCA2 mutations correlate with TP53 ab-normalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol 2012;25:740-50. 10.1038/modpathol.2011.211 [DOI] [PubMed] [Google Scholar]
- 77.Oza AM, Cibula D, Benzaquen AO. Olaparib combined with chemotherapy for recurrent plati-num-sensitive ovarian cancer: a randomised phase 2 trial (vol 16, pg 87, 2015). Lancet Oncol 2015;16:E55. 10.1016/S1470-2045(14)71135-0 [DOI] [PubMed] [Google Scholar]
- 78.Bang YJ, Im SA, Lee KW, et al. Randomized, Double-Blind Phase II Trial With Prospective Classi-fication by ATM Protein Level to Evaluate the Efficacy and Tolerability of Olaparib Plus Paclitaxel in Patients With Recurrent or Metastatic Gastric Cancer. J Clin Oncol 2015;33:3858-65. 10.1200/JCO.2014.60.0320 [DOI] [PubMed] [Google Scholar]
- 79.Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immuno-therapy: more haste, less speed. Br J Cancer 2018;118:312-24. 10.1038/bjc.2017.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiao S, Xia W, Yamaguchi H, et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin Cancer Res 2017;23:3711-20. 10.1158/1078-0432.CCR-16-3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barker HE, Patel R, McLaughlin M, et al. CHK1 Inhibition Radiosensitizes Head and Neck Cancers to Paclitaxel-Based Chemoradiotherapy. Mol Cancer Ther 2016;15:2042-54. 10.1158/1535-7163.MCT-15-0998 [DOI] [PubMed] [Google Scholar]
- 82.Hafsi H, Dillon MT, Barker HE, et al. Combined ATR and DNA-PK Inhibition Radiosensitizes Tumor Cells Independently of Their p53 Status. Front Oncol 2018;8:245. 10.3389/fonc.2018.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLaughlin M, Barker HE, Khan AA, et al. HSP90 inhibition sensitizes head and neck cancer to platin-based chemoradiotherapy by modulation of the DNA damage response resulting in chromosomal fragmentation. BMC Cancer 2017;17:86. 10.1186/s12885-017-3084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dillon MT, Bergerhoff KF, Pedersen M, et al. ATR Inhibition Potentiates the Radiation-induced Inflammatory Tumor Microenvironment. Clin Cancer Res 2019;25:3392-403. 10.1158/1078-0432.CCR-18-1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schiewer MJ, Knudsen KE. Linking DNA Damage and Hormone Signaling Pathways in Cancer. Trends Endocrinol Metab 2016;27:216-25. 10.1016/j.tem.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brenner JC, Ateeq B, Li Y, et al. Mechanistic Rationale for Inhibition of Poly(ADP-Ribose) Polymerase in ETS Gene Fusion-Positive Prostate Cancer (vol 19, pg 664, 2011). Cancer Cell 2013;23:557. 10.1016/j.ccr.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schiewer MJ, Goodwin JF, Han S, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov 2012;2:1134-49. 10.1158/2159-8290.CD-12-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muvarak NE, Chowdhury K, Xia L, et al. Enhancing the Cytotoxic Effects of PARP Inhibitors with DNA Demethylating Agents - A Potential Therapy for Cancer. Cancer Cell 2016;30:637-50. 10.1016/j.ccell.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pulliam N, Fang F, Ozes AR, et al. An Effective Epigenetic-PARP Inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA Mutations. Clin Cancer Res 2018;24:3163-75. 10.1158/1078-0432.CCR-18-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Criscuolo D, Morra F, Giannella R, et al. New combinatorial strategies to improve the PARP inhibitors efficacy in the urothelial bladder Cancer treatment. J Exp Clin Cancer Res 2019;38:91. 10.1186/s13046-019-1089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as