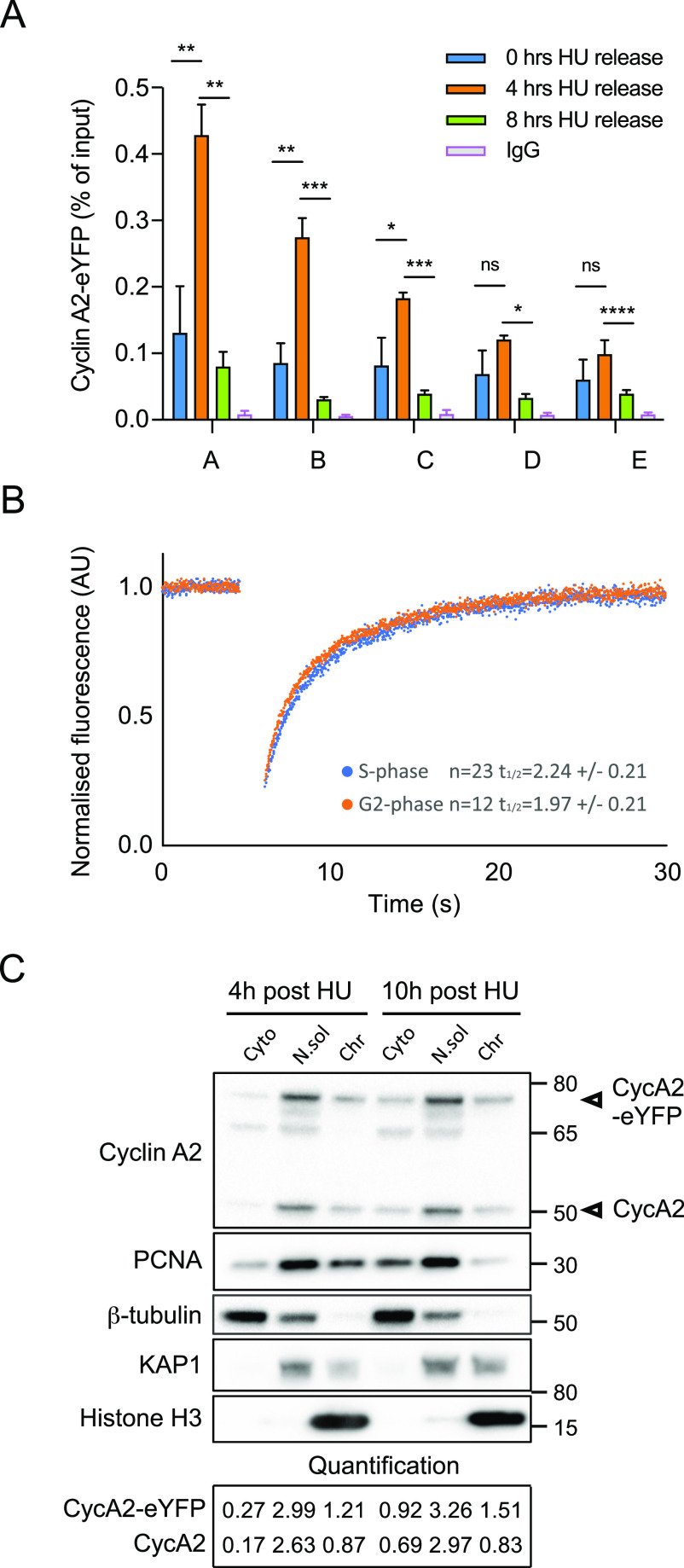
Figure 5. Cyclin A2 associates with chromatin during the S phase.
(A) Chromatin immunoprecipitation (ChIP) in RPE-CycA2-eYFP cells, synchronised with hydroxyurea (HU, 2 mM in PBS) for 18 h and harvested after HU washout at different time points (0, 4 and 8 h). The enrichment of CycA2-eYFP and IgG, determined by ChIP-qPCR is plotted as percentage of input (ratio of immunoprecipitated DNA to the total amount of DNA). Genomic locations are indicated by alphabetical letters A–E (details in Table S1). Graph shows one representative experiment. Mean value and SD are based on duplicate samples, each with duplicate qPCR reactions. Mean relative recovery for non-immune IgG are 0.0072. Asterisks indicate two-tailed paired t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) FRAP in the nucleus of RPE-CycA2-eYFP cells. Graph shows normalised average of cells with (G2 phase) and without (S phase) detectable cytoplasmic CycA2-eYFP. N = 2. (C) RPE-CycA2-eYFP cells were synchronised with hydroxyurea for 18 h and harvested after 4 and 10 h in HU-free media. Cytoplasmic proteins (Cyto), soluble nuclear proteins (N.sol), and chromatin bound proteins (Chr) were isolated and detection of Cyclin A2 and PCNA was performed by Western blot. 5 μg of protein was loaded for each fraction. β-Tubulin, KAP1, and Histone H3 were used as controls of cytosolic, nuclear soluble and chromatin fraction, respectively. N = 2.