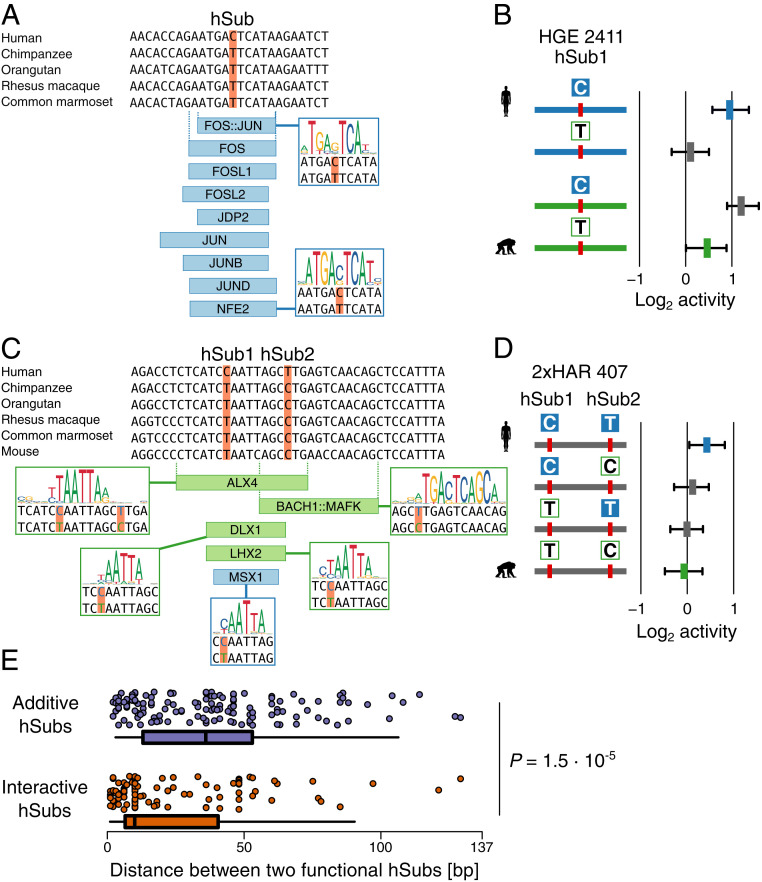
Fig. 6.
Changes in predicted transcription factor binding sites due to hSubs that alter enhancer activity. (A) Five-primate alignment over an hSub in HGE 2411. The alignment was derived from the 100-way Multiz alignment (GRCh37/hg19, University of Santa Cruz Genome Browser; http://genome.ucsc.edu) . Nine TFs are predicted to bind the human, but not the chimpanzee ortholog (TFBSs predicted to show increases in affinity are highlighted in blue). (B) MPRA activity of the hSub shown in A, as well as the activities of the chimpanzee ortholog and synthetic intermediates. The figure is labeled as in Fig. 4A. (C) Alignment of a pair of hSubs in 2xHAR 407 that overlaps a cluster of TFBSs (blue, stronger predicted binding in human; green, stronger predicted binding in chimpanzee). See SI Appendix, Fig. S9 for additional TFBSs in this locus that are not affected by the hSubs. (D) MPRA activity of the two hSubs shown in C and the corresponding chimpanzee ortholog and synthetic intermediates. (E) Distribution of physical distances between pairs of additive (Top, in blue) and interactive (Bottom, in orange) hSubs with regulatory effects. BH-corrected P values were calculated using a Mann–Whitney U test.