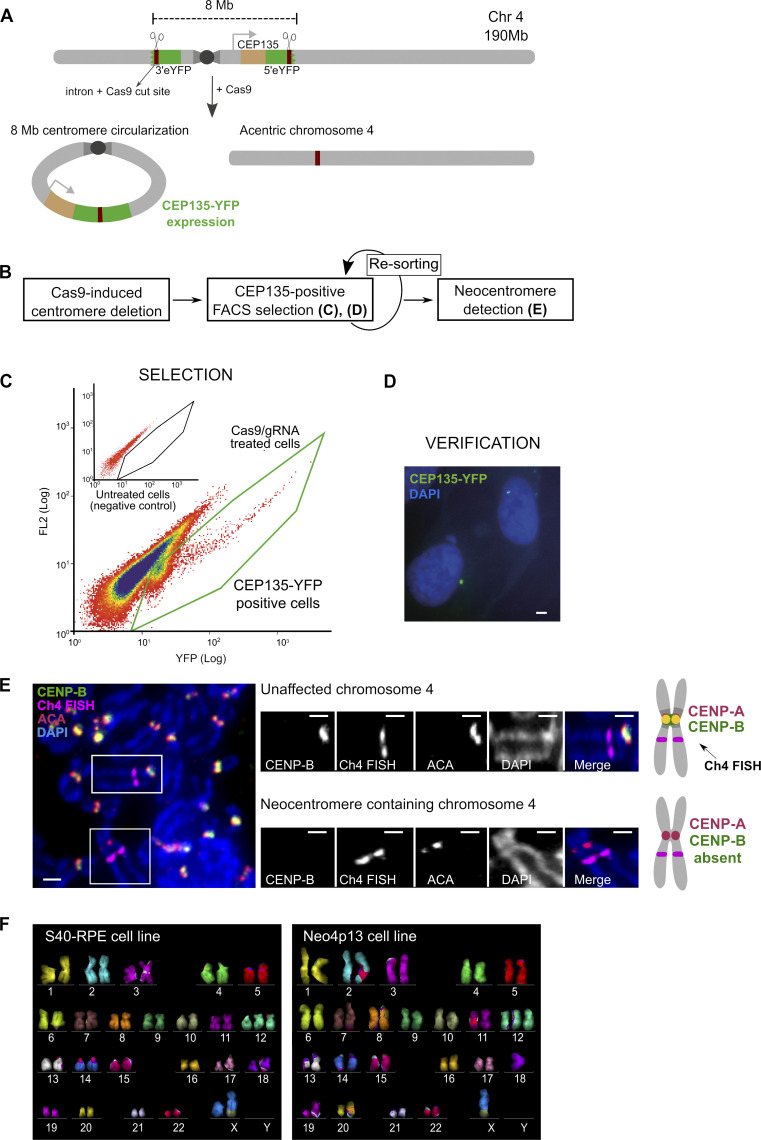
Figure 1.
Strategy for human centromere deletion and neocentromere isolation. (A) A cassette carrying the 5′ portion of the eYFP was integrated on the 4q-arm in frame with CEP135 protein (light brown) and the 3′ portion on the 4p-arm, flanking the centromere. Cassettes carry a murine intron (dark brown) that acts as a gRNA target for Cas9. Upon Cas9 cleavage one possible outcome is the circularization of the centromere fragment (∼8 Mb) leading to reconstitution of spliced CEP135-eYFP. The acentric chromosome serves as template for neocentromere formation (see Fig. S1). (B) Experimental design for centromere deletion and neocentromere selection and detection. (C) Selection of CEP135-eYFP positive cells by flow cytometry (FL2, fluorescence channel 2 used for scatter and autofluorescence measurements). (D) Micrograph showing CEP135-eYFP foci indicative of centromere fragment circularization. Scale bar, 2 µm. (E) Micrograph of neocentromere detection by FISH-IF in mitotic spreads. The FISH probe identifies chromosome 4, ACA (CENP-A, CENP-B, and CENP-C) localizes to active centromeres, and CENP-B binds specifically to alphoid DNA, absent from neocentromeres. Insets display the two chromosome 4s from the image on the left. Scale bars, 2 µm. Diagram on the right represents the expected staining pattern to identify the neocentromere. (F) Karyotypes of the parental and neocentromere cell lines by mFISH. S40-RPE carries a preexisting translocation (X,10) and an extra copy of chromosome 12, present in all the clones analyzed. Chr4, chromosome 4.