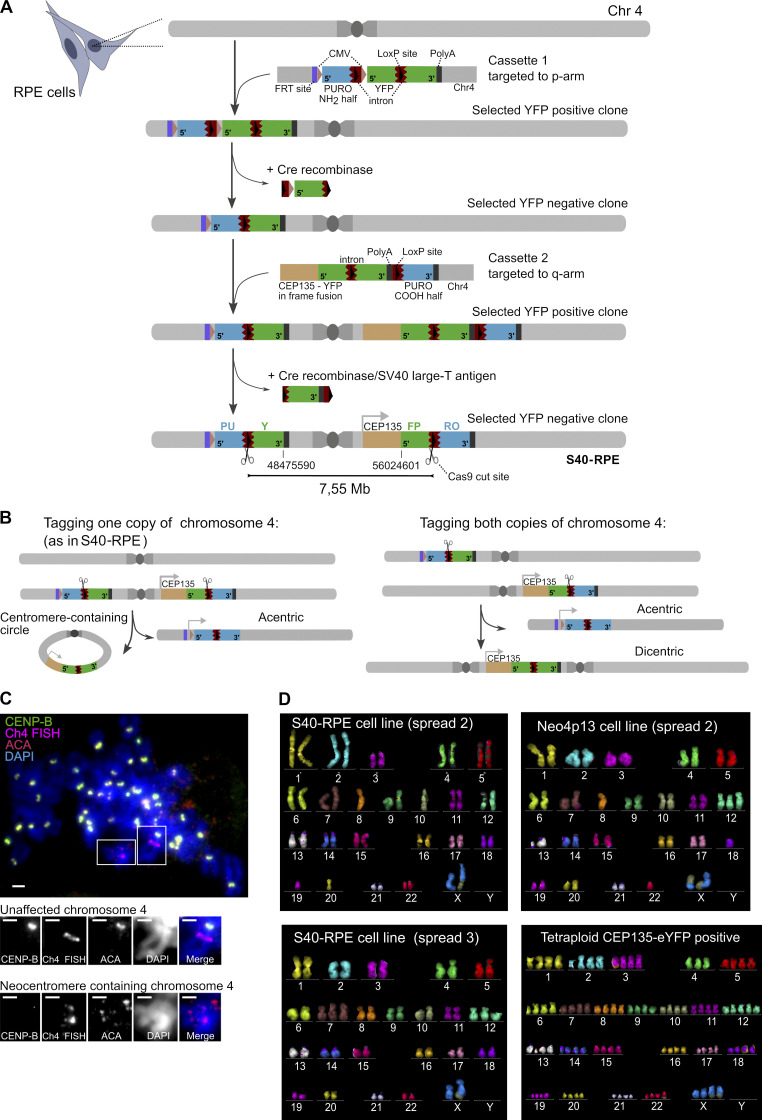
Figure S1.
Genomic architecture of strategy for centromere deletion and karyotype of neocentromere-containing cells and alternative survivors. (A) Scheme of the sequential targeting steps and details of the targeting cassettes inserted into RPE cells flanking the centromere of chromosome 4. Constructs carry 5′ and 3′ fragments of the eYFP (green) split across the two genomic locations, separated by a mouse intronic sequence (brown) that is spliced efficiently (Lacy-Hulbert et al., 2001), and act as a target for gRNAs that will recruit Cas9 nuclease (scissors). Introns also carry a loxP site (black arrowhead). 5′ and 3′ fragments of a split Puromycin resistance gene (blue) are positioned downstream of the introns. Note that for the purposes of targeting the cassettes, a full-length eYFP gene was used to aid in fluorescence-based selection of targeted clones, followed by Cre-mediated removal of indicated gene fragments, ultimately resulting in the split eYFP arrangement flanking the centromere. Following Cas9 cut and repair, one possible outcome is the circularization of the centromere fragment (7.55-Mb region), leading to reconstitution of eYFP in frame with the centriole-associated protein CEP135. (Note that imperfect repair following the Cas9 cut is tolerated within the intronic region.) Conversely, the fusion of the acentric arms of the chromosome will lead to reconstitution of the Puromycin resistance gene driven from a cytomegalovirus (CMV) promoter (pink arrowhead) not used in this study. On the p arm, we also included an FRT (Flippase Recognition target) recombination site (purple), not used in this study. Indicated coordinates correspond to GRCh38 assembly. (B) Targeting of the p- and q-arm constructs from (A) result in two possible allele arrangements. Both possible scenarios (p- and q-targeted cassettes both inserted into the same chromosome 4 homologue [1] or in different ones [2]) lead to an acentric chromosome 4 formation after the Cas9 cut, serving as template for neocentromere formation. Based on mitotic spreads, multicolor karyotyping, and sequencing data, we deduced the S40-RPE parental cell line used in this study is of the scenario 1 type. (C) Early neocentromere detection by FISH-IF (performed as in Fig. 1 E) on the first CEP135-YFP–positive polyclonal population sorted 72 h after Cas9 electroporation and analyzed 3 d later. Scale bars, 2 µm. (D) Karyotype of indicated cell lines based on mFISH on different spreads, shortly after FACS-based isolation. Translocations and aneuploidies are detected in both the parental S40-RPE and Neo4p13 cell lines, possibly driven by SV40 expression. The tetraploid karyotype of a CEP135-eYFP–positive clone without an acquired neocentromere is also shown, indicative of single chromosome 4 loss. Chr, chromosome.