Abstract
Correct cellular localization is essential for the function of many eukaryotic proteins and hence cell physiology. Here, we present a synthetic genetic device that allows the control of nuclear and cytosolic localization based on controlled alternative splicing in human cells. The device is based on the fact that an alternative 3′ splice site is located within a TetR aptamer that in turn is positioned between the branch point and the canonical splice site. The novel splice site is only recognized when the TetR repressor is bound. Addition of doxycycline prevents TetR aptamer binding and leads to recognition of the canonical 3′ splice site. It is thus possible to produce two independent splice isoforms. Since the terminal loop of the aptamer may be replaced with any sequence of choice, one of the two isoforms may be extended by the respective sequence of choice depending on the presence of doxycycline. In a proof-of-concept study, we fused a nuclear localization sequence to a cytosolic target protein, thus directing the protein into the nucleus. However, the system is not limited to the control of nuclear localization. In principle, any target sequence can be integrated into the aptamer, allowing not only the production of a variety of different isoforms on demand, but also to study the function of mislocalized proteins. Moreover, it also provides a valuable tool for investigating the mechanism of alternative splicing in human cells.
Keywords: aptamer, TetR, 3′ splice site, alternative splicing, nuclear import
INTRODUCTION
Eukaryotic cells are separated into different morphological and functional compartments to efficiently carry out highly diverse processes. For example, the spatial separation of replication and transcription in the cell nucleus and protein translation in the cytoplasm requires a strictly regulated network to ensure the targeted transport of macromolecules required for these processes into and out of the cell nucleus. Evidently, correct transport processes are essential for cell physiology. Aberrant spatiotemporal localization of proteins caused by mutation, altered expression of cargo proteins or transport receptors or deregulation of components of the trafficking machinery have been linked to the onset of disease, tumorigenesis and metastasis (Hung and Link 2011; Hill et al. 2014). Despite its importance, very few inhibitors that interfere with nuclear transport have been identified to date. Due to the fact that the nucleocytoplasmic transport machinery is rather generic, pharmacological strategies currently available are limited (Kim et al. 2017; Jans et al. 2019). Hence the development of synthetic genetic devices enabling spatiotemporal regulation of transport processes is of great benefit for phenotypic studies and therapeutic strategies that target protein localization (Hung and Link 2011; Niopek et al. 2016).
Active nucleocytoplasmic shuttling is mediated by short genetically encoded sequences, so-called nuclear localization/export sequences (NLS/NES) that determine the localization of a protein. Synthetic devices that control the accessibility of these sequences can thus be applied to pinpoint a protein to a specific location. Optogenetic tools have been developed for light-induced nuclear import and export (Niopek et al. 2014, 2016). For these tools, the import/export sequences are concealed within a photosensitive protein domain that unfolds upon illumination. In contrast, we propose an alternative approach to control the availability of the NLS by means of alternative splicing.
Alternative splicing is a crucial cellular process responsible for the diversity of the human proteome and the coordination of physiological functions in different developmental processes (Baralle and Giudice 2017). The accuracy of the splicing process involves the recognition of short sequences within the pre-mRNA, that is, the 5′ and 3′ splice sites (SS), that delimit the exon–intron boundaries (Will and Lührmann 2011; Vigevani et al. 2017). In introns removed by the major (U2-dependent) spliceosome, the 3′SS is encoded mostly by the dinucleotide AG, which is present in the genome very frequently. In mammalian genes the mechanism of 3′SS selection is complex and still not fully understood (Chen et al. 2017). The spliceosome is assisted in the 3′SS identification by a polypyrimidine (Py)-tract, a sequence located between the branch point sequence (BP) and the AG dinucleotide (Guth et al. 1999; Horowitz 2012; Pérez-Valle and Vilardell 2012). The Py-tract is recognized by U2AF65, which acts in a complex with the splicing factor SF1 and the spliceosomal subunit U2AF35 that recognize the upstream BP and the consensus AG dinucleotide at the 3′SS, respectively. Moreover, it has been demonstrated that AG selection depends on the BP-to-AG and AG-to-AG distances (Chua and Reed 1999, 2001). All of this implies flexibility in the recognition of 3′SS, BP and Py-tract sequences and thus the possibility of alternative splicing in mammalian introns. In the present study, we explored the targeted control of alternative splicing by using an aptamer.
Our aptamer of choice was the TetR-binding RNA aptamer identified by a combination of in vitro selection for TetR binding and an in vivo screening for its ability to activate TetR controlled transcription (Hunsicker et al. 2009). It folds into a stem–loop structure with an internal asymmetric loop that displays the protein-binding site (Fig. 1A; Hunsicker et al. 2009). The recently resolved crystal structure of the RNA aptamer-TetR-complex shows that the aptamer recognizes the helix-turn-helix motif of TetR and consequently the same binding site that is recognized by the operator DNA (Fig. 2A; Grau et al. 2020). We were fortunate to happen to find that the occurring conformational changes of TetR upon tetracycline binding prevent not only operator DNA but also RNA aptamer binding, and in equal measures (Tiebel et al. 1999; Steber et al. 2011). Consequently, this allows reversible tetracycline-dependent binding of TetR to both DNA and RNA and thus opens up a wide range of possible applications of the TetR aptamer as genetic device. While the original publication focused on the control of transcription in Escherichia coli (Hunsicker et al. 2009), portability and broader applicability of the system was documented with its successful use in the protozoon Plasmodium falciparum and in yeast (Goldfless et al. 2012; Ganesan et al. 2016). The TetR aptamer was then applied to control miRNA biogenesis in human cells (Atanasov et al. 2017). In this specific approach, the TetR aptamer replaces the natural terminal loop of precursor miRNAs, which, upon binding of TetR, leads to the inhibition of miRNA processing by Dicer via steric hindrance. The inhibition is fully reversible after addition of doxycycline, thus providing a system that allows control over intracellular miRNA levels and, consequently, their gene-silencing properties. The latest approach (Mol et al. 2019) exploits the TetR-binding aptamer for the control of translation initiation and pre-mRNA splicing. For this, the TetR aptamer was inserted into the 5′ untranslated region in such a way that it interferes with ribosomal scanning when bound by TetR. Moreover, upon insertion in proximity to the 5′SS, intron retention was controlled via the binding of TetR to the aptamer. Again, in both approaches TetR binding is fully relieved by the addition of doxycycline (Mol et al. 2019).
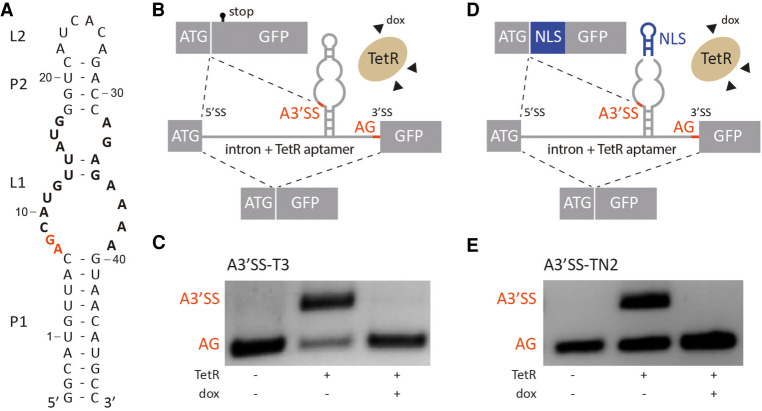
FIGURE 1.
TetR aptamer controls alternative 3′ splice site recognition. (A) Secondary structure of the TetR aptamer. Stems and loops are indicated with P and L, respectively. Nucleotides involved in TetR binding are highlighted in bold, the A3′SS A7G8 is labeled in red. (B,D) Schematic of the proposed model. In the absence of TetR, canonical AG is recognized by the spliceosome. When TetR binds to the aptamer, the A3′SS is activated. The splicing pattern can be restored with the addition of doxycycline (dox) that leads to conformational changes of TetR and the release of the aptamer. Controlled production of two splice variants is possible with this mechanism. Additionally, insertion of a nuclear localization sequence (NLS, in blue) between A3′SS and AG (D) allows to produce different splice variants with different subcellular localization on demand. Exons are displayed as boxes and the intron with the TetR aptamer is shown as a line. (C,E) Splicing pattern visualized by RT-PCR. HeLa cells were transiently transfected with the constructs A3′SS-T3 and A3′SS-TN2 and cotransfected with plasmid expressing TetR (+) and treated with (+) or without (−) 50 µM doxycycline (dox) for 24 h. Total RNA was prepared and used for RT-PCR with primer pairs binding to both exons. The upper band corresponds to usage of A3′SS and the lower band to distal AG. The experiment was repeated three times with similar results.
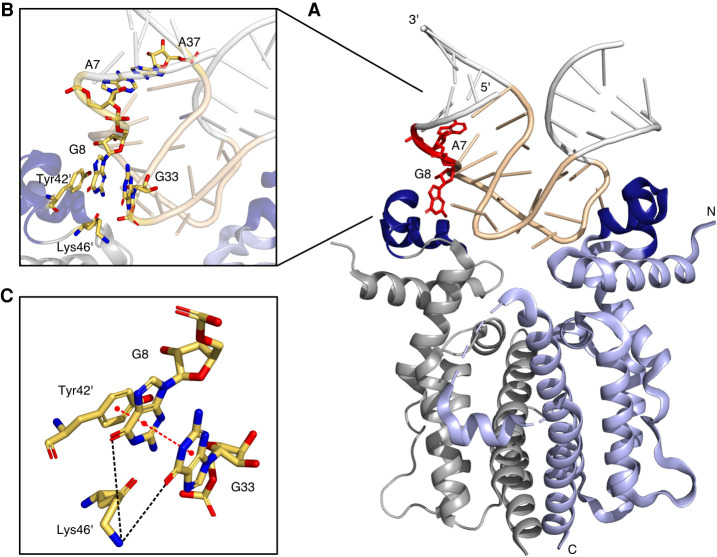
FIGURE 2.
Crystal structure of the RNA aptamer-TetR-complex (PDB 6SY4, Grau et al. 2020). (A) Cartoon representation of dimeric TetR (in blue and gray) in complex with the TetR aptamer. Amino- and carboxyl termini and 5′ and 3′ ends are labeled, helix-turn-helix motifs formed by the helices α2 and α3 of each monomer are colored in dark blue. Stick representation of the nucleotides A7 and G8 are displayed in red. (B) Close up view of the TetR aptamer interface. (C) Stick representation of G8, G33, Tyr42′, and Lys46′.
In the present study, we further extended the application spectrum of the TetR aptamer as a synthetic device. We engineered a system that allows us to activate an alternative 3′SS. Depending on whether TetR is bound or not, the canonical or an alternative 3′SS (A3′SS) is used, thus producing two splice variants. If the sequence of a specific protein domain is then inserted between the two 3′SS, it becomes a matter of deliberate choice whether this protein domain is incorporated into the protein or not. Moreover, we have successfully demonstrated the proof-of-concept for a nuclear localization sequence by transporting a 110 kDa cytoplasmic GFP-fusion protein into the nucleus, depending on doxycycline. Our results promise a robust and reliable option for a wide range of applications.
RESULTS
As a first step toward creating a device for the control of alternative 3′ splicing, the MINX intron with flanking exon sequences was placed in front of a GFP reporter gene. The small intron derives from the adenovirus genome and is efficiently spliced in human cells (Supplemental Fig. S1; Zillmann et al. 1988). The TetR aptamer (Fig. 1A) was then placed upstream of the canonical 3′SS (Fig. 1B). It is important to mention that the TetR aptamer itself contains several potential sequences that can be recognized as A3′SS and we expected one of these to be used. The splicing patterns of the constructs were analyzed in transiently transfected HeLa cells in the absence and presence of doxycycline. A plasmid expressing TetR equipped with a NLS was cotransfected. The splicing pattern was visualized by RT-PCR using primer pairs binding to both exons (Fig. 1C). In the absence of TetR, solely the canonical 3′SS was used. However, usage of an A3′SS occurred when TetR was coexpressed (see Fig. 1C, middle lane). Interestingly, the sequence analysis of the splice products confirmed that exclusively the first A3′SS of the aptamer (A7G8) was activated and primarily used in comparison with the canonical 3′SS (Supplemental Fig. S2). Other potential A3′SS were not used at all. After addition of doxycycline and the resulting detachment of TetR from the aptamer, usage of the canonical 3′SS was restored.
Since RNA-based synthetic devices are highly dependent on genetic context and stability of the regulatory RNA element (Vogel et al. 2018; Groher et al. 2019; Mol et al. 2019), the TetR aptamer was optimized for both the lengths of the closing stem P1 and of the Py-tract (Supplemental Fig. S3; B Suess, unpubl.), resulting in A3′SS-T3 as the construct with the best switching behavior. qRT-PCR was performed to quantify the observed effect on regulation (Supplemental Fig. S4). A mutation in the aptamer that suspends TetR binding completely prevented alternative splicing (A3′SS-T3*, Supplemental Fig. S3F), thus demonstrating that the observed effect is in fact caused by TetR binding. In sum, we have developed a system that can efficiently and reliably switch between two splice isoforms dependent on the ligand doxycycline.
Inspired by the cell cycle-regulated Nek2 kinase (Wu et al. 2007) that controls its cellular localization by alternative splicing, we aimed to apply our system to conditionally control localization. For this purpose, a localization sequence had to be inserted between the two 3′SS. Fortunately, the upper stem–loop of the aptamer (P2/L2) is not relevant for TetR binding and can therefore be modified as long as the stability of stem P2 is preserved (Hunsicker et al. 2009). This allows the insertion of any new sequence including, for example, an NLS. Consequently, we exchanged a part of the stem–loop with the sequence of the c-MYC NLS (Fig. 1D; Supplemental Fig. S5) resulting in the constructs A3′SS-TN1 and -TN2 that only differ in the length of the closing stem P1. In addition, we carefully adjusted the distance between aptamer and 3′SS to maintain NLS and GFP in an open reading frame (Supplemental Fig. S5). The construct A3′SS-TN2 proved to be the construct with the best splicing behavior with similar efficiency as the parental construct A3′SS-T3 (compare Fig. 1C with 1E). qRT-PCR was performed to quantify the observed effect on regulation (Supplemental Fig. S4) and confirmed that the insertion of the NLS does not influence the switching properties. Sequencing analyzes verified that the resulting splice variants differ in the presence or absence of an NLS.
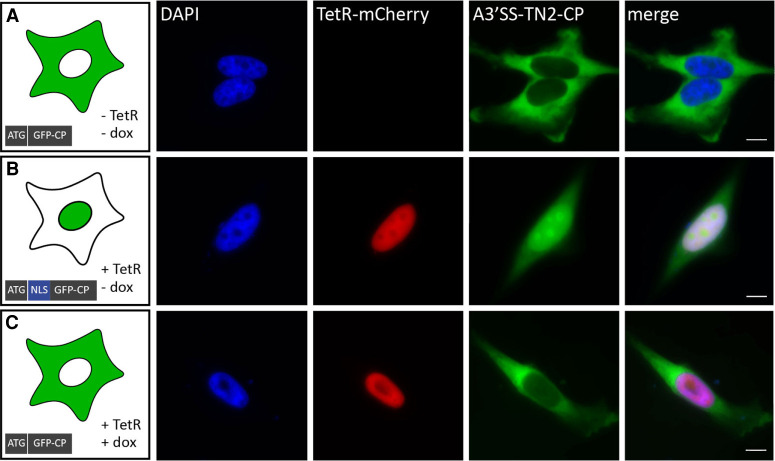
Next, we examined the potential of the construct A3′SS-TN2 for controlled cellular localization. To avoid passive diffusion of the rather small GFP (Supplemental Fig. S6), we fused it to a larger cytosolic protein (CP) resulting in the construct A3′SS-TN2-CP. The resulting fusion protein GFP-CP (about 110 kDa) was expected to be primarily located within the cytoplasm. As cytosolic protein we used SERPINE1 mRNA-binding protein 1 (SERBP1), an RNA-binding protein with predominantly cytoplasmic localization. We stably integrated A3′SS-TN2-CP into the HeLa cell line HF1-3 using the Flp-In system. The generated cell line expressing A3′SS-TN2-CP was subsequently transiently transfected with a TetR-mCherry expressing plasmid. After transfection, cells were treated with or without 50 µM doxycycline for 24 h, then fixed, stained with DAPI and analyzed for both GFP and mCherry expression. The microscopic images of a control cell carrying only GFP showed a homogenous distribution throughout the cell with a tendency to accumulate in the nucleus due to passive diffusion (Supplemental Fig. S6). Constitutively expressed A3′SS-TN2-CP showed predominant cytoplasmic localization (Fig. 3A), thus confirming that only the canonical 3′SS was used in the production of the isoform without NLS. In the presence of TetR, GFP-CP expression in the cytosol decreased and GFP-CP accumulated primarily in the nucleus (Fig. 3B). This indicates that the A3′SS was activated and the alternative splice isoform carrying the NLS was produced. Additionally, cells expressing TetR and incubated with doxycycline showed mostly cytoplasmic distribution (Fig. 3C), indicating that doxycycline lead to the release of TetR from the pre-mRNA.
FIGURE 3.
Microscopic visualization of inducible control of nuclear import with a TetR aptamer. (A) A3′SS-TN2-CP stably integrated into an HF1-3 cell line using the Flp-In system. (B,C) The generated cell line expressing the A3′SS-TN2-CP construct was transiently transfected with mCherry-tagged TetR (+) and treated with (+) or without (−) 50 µM doxycycline (dox) for 24 h. Cells were fixed and stained with DAPI. Scale bar, 10 µm.
DISCUSSION
In this study, we succeeded in the development of a synthetic control device that allows the deliberate and reversible activation of an alternative 3′SS. It is therefore possible to selectively produce two independent splice isoforms. We have used the system to attach a specific transport sequence to one splice isoform and thus directed a cytosolic protein into the nucleus. In consequence, our system represents an alternative to the recently developed optogenetic tools that also allow a targeted nuclear transport (Niopek et al. 2014, 2016). These systems, however, are limited to proteins that will tolerate the attachment of a photosensitive domain. Where this is not feasible, our system may offer a convenient alternative since there is only the NLS attached to the target protein and no major modifications are necessary. Moreover, our system is not limited to nuclear import; in principle, any sequence can be attached to the isoform. Thus, in addition to the application demonstrated here, the system technically allows the targeted transport of proteins to any compartment for which transport tags have been described. Moreover, it is also possible to switch between different splice isoforms in a doxycycline-dependent manner to study their function.
Although the system described here is very reliable, the underlying mechanism has not yet been fully understood in detail. Our initial assumption was that one or more of the potential 3′ SS of the TetR aptamer are used and its recognition can then be blocked by TetR. A similar approach has been described using a theophylline aptamer (Kim et al. 2005, 2008; Gusti et al. 2008). Here, a proximal 3′SS was embedded within the theophylline aptamer that was—in contrast to our system—preferentially used. Addition of theophylline then promoted splicing at the distal 3′SS, thus changing the ratio of distal-to-proximal 3′SS usage by about twofold. In our approach, however, we observed the exact opposite behavior. In the absence of TetR, only the canonical/distal 3′SS is recognized while all alternative/proximal 3′SSs are sequestered in the aptamer structure. The results are in line with an analysis for 3′SS selection in Saccharomyces cerevisiae that examines the importance of RNA secondary structures within the intronic region between the BP and the 3′SS. Here, the usage of the 3′SS is higher if it is not concealed within a stem structure (Meyer et al. 2011).
In mammalian introns, the Py-tract plays an essential role in the splicing process. It acts at an early stage of spliceosome assembly and is required for the first step of the catalytic process. The length of the Py-tract and its composition are important in 3′SS recognition (Hallegger et al. 2010; Jenkins et al. 2013). We also observed this. The location of the Py-tract relative to the aptamer position as well as their nucleotide composition highly influences the selection of the 3′SS and significantly affects the switching properties (B Suess, unpubl.). Interestingly, the presence of the aptamer structure between a Py-tract and the 3′SS does not interfere with the splicing process. Instead, the aptamer appears to be required to reduce the distance between the BP and the 3′SS. The Py-tract can also be divided by the aptamer (see A3′SS-T1; Supplemental Fig. S3). Presumably, the Py-tract sequences are merged here by the aptamer to facilitate the recognition of splicing factors. However, unexpectedly, an A3′SS is activated upon TetR binding to the aptamer. A possible explanation here could be that the second step of splicing may be blocked by interference of the RNA aptamer-TetR-complex with the SF1–U2AF65 and U2AF65–U2AF35 binding to the canonical 3′SS. The accumulation of splicing factors next to the aptamer may then lead to A3′SS recognition.
Alternatively, TetR binding may open up the binding site and thus in some way expose the CAG sequence. If at all, the crystal structure could explain this only for the first of the three possible alternative splice sites (A7G8, see Figs. 1A, 2). The second and third A3′SS are tightly embedded into the aptamer structure as part of the terminal loop L2 (A27G28) and A32G33 as part of a purine stack. The first A3′SS is located directly next to the closing stem P1, which is flanked by a noncanonical base pair classified as A7:A39 (A37crystal) trans Watson-Crick/Watson-Crick (Fig. 2, please note: A39 in Figure 1A corresponds to A37 in the crystal structure due to a different loop L2 size used for crystallization). A7 is stacking on the neighboring base of the helix P1, whereas G8 is flipped outwards and points toward the protein with torsion angles adopting nonstandard values (Fig. 2B). However, the flipped-out base G8 stacks directly with Tyr42′ of TetR. There is also an additional layer of π-π stacking between G8 and the likewise flipped G35 (G33crystal, Fig. 2C). In addition, the ε-amino group of Lys46′ is located within hydrogen-bonding distance of both G8 and G35 (G33crystal). However, it was not possible to accurately determine the exact orientation of the side chain in the crystal structure due to a lack of well-defined electron density. This indicates that the hydrogen bond is only formed in some molecules in the crystals and that, overall, this hydrogen bond does not contribute significantly to the free binding energy of the complex formation. Consequently, a certain flexibility in this region may be assumed. With both bases not enclosed in a helical region and G8 actually flipped out, it could explain why the splice site may be recognized in the presence of TetR. So far, we do not know the shape of the aptamer in the absence of the protein. In inline probing, we only see very weak signals for both positions indicating a low flexibility (Hunsicker et al. 2009). For instance, for the flipped-out G8, a stronger signal should be expected from this assay. This might indicate that the aptamer in the free form may adopt a different conformation that prevents splice site recognition. For the second and third A3′SS, no protein-dependent changes could be detected by inline probing. However, to finally resolve this, the structure of the free aptamer would have to be determined including the dynamics of the individual bases.
In sum, we developed a system that allows ligand-dependent activation of an alternative 3′SS. In a proof-of-concept study, we were able to fuse a transport tag to a cytosolic target protein, thereby directing it into the nucleus. However, our system is not limited to the control of nuclear localization. In principle, any target sequence can be integrated into the aptamer, allowing not only the production of a variety of different isoforms on request, but also to study the function of mislocalized proteins. Moreover, it also provides a valuable tool for investigating the mechanism of alternative splicing in human cells.
MATERIALS AND METHODS
Cell culture
HeLa cells (DSMZ, no. ACC-57) and HF1-3 cells “Flp-In Host Cell Line” (Berens et al. 2006) were maintained at 37°C in a 5% CO2 humidified incubator and cultured in Dulbecco's Modified Eagle Medium (DMEM, Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS Superior, Biochrom), 100 U/mL penicillin (PAA, the Cell Culture Company), 100 µg/mL streptomycin (PAA, the Cell Culture Company) and 1 mM sodium pyruvate (PAA, The Cell Culture Company). For HF1-3 cells, 150 μg/mL zeocin (Invitrogen) was additionally supplemented to the medium, whereas the medium of the HF1-3 cells with the integrated constructs GFP control and A3′SS-TN2-CP contained 200 μg/mL hygromycin (Invitrogen).
Plasmid construction
Plasmids were constructed by standard cloning techniques using overlap extension PCR with Q5 Polymerase (NEB) and restriction and ligation reactions with HF restriction enzymes (NEB) and T4-DNA ligase (NEB), respectively. All vector maps and PCR primers are available upon request. Custom oligonucleotides were synthesized by Sigma Aldrich. All constructs were cloned into pcDNA5/FRT vector under the CMV promoter (Mol et al. 2019). TetR protein was expressed from CMV promoter and modified at the N-terminus with a nuclear localization signal from c-MYC (5′-CCGGCCGCGAAACGCGTGAAACTGGAT-3′). The construct expressing TetR-mCherry was cloned by insertion of the mCherry gene by unique restriction sites for KpnI and AgeI.
Genomic integration
HF1-3 cells were transfected with the plasmids pcDNA5/FRT and pOG44 (recombinase expression plasmid, Invitrogen) at a molar ratio of 1:9 using Lipofectamine 2000 (Life Technologies) according to manufacturer instructions. The medium was changed 24 h after transfection to DMEM. The cells were selected for stable integration by adding 200 µg/mL hygromycin. After 2 wk of selection with hygromycin, cells were sorted by the S3e Cell Sorter (Bio-Rad) for GFP positive cells and analyzed for stable integration by genomic PCR and sequencing.
RT-PCR
A total of 120,000 HeLa cells per well were seeded into a 12-well plate. The cells were transfected with 100 ng reporter DNA and 300 ng TetR plasmid using 2 µL of Lipofectamine 2000 (Life Technologies) per well according to manufacturer instructions. Then, 2 h after transfection, the medium was changed to DMEM with or without 50 µM doxycycline, and the cells were incubated at 37°C and 5% CO2 for 24 h. RNA was isolated using TRIzol (Life Technologies) according to the manufacturer's protocol. Contaminating DNA was removed with the TURBO DNA-free kit (Life Technologies), and the RNA was quality-checked on a 1% (w/v) agarose gel. Next, 1 µg RNA was reverse-transcribed by MuLV (Applied Biosystems) using random hexamers (Fermentas) with the supplied buffers (10 min at 20°C, 15 min at 42°C, 5 min at 99°C). Then, 50 ng cDNA was PCR amplified using Taq polymerase (New England Biolabs, initial denaturation 2 min at 96°C, 30 sec at 96°C, 30 sec at 54°C, 30 sec at 72°C, 35 cycles) and analyzed on a 3% (w/v) agarose gel. The amplified products were cloned (CloneJET PCR Cloning Kit, Thermo Scientific) and sequenced for verification. Each RT-PCR was repeated in three independent experiments
Primers are listed below.
Exon-1-f AACCCGTCGGCCTCCGAAC
Exon-2-r CGCCGGACACGCTGAACTTG
qPCR analysis
For qPCR analysis, the Fast SYBR Green Master Mix (Applied Biosystems) was used and the samples were analyzed on a StepOnePlus Real-Time PCR machine (Applied Biosystems). The total volume of 20 μL of a standard mix for one reaction was 10 μL SYBR Green Master Mix (2×), 5 μL cDNA and 1 μL of a primer mix of the respective two gene-specific primers (10 μM). The PCR program for SYBR Green-based qPCR was 20 sec at 95°C, 3 sec at 95°C, 30 sec at 60°C, 40 cycles and for the melting curve: 15 sec at 95°C, 60 sec at 60°C, 15 sec at 95°C. Primers are listed below. Analyses were performed with samples from three independent experiments in technical replicates. Results were calculated according to the ΔΔCt method (Pfaffl 2001).
pre-mRNA-f CGGCCTCCGAACGATGGTAAGAGCC
A3′SS-f CGGCCTCCGAACGATGCATGTTATG
3′SS-f CGGCCTCCGAACGATGCTCGCGGTT
GFP-r TCGCCGGACACGCTGAACTTG
Actin-f CGGGACCTGACTGACTACCTC
Actin-r CTTCTCCTTAATGTCACGCACG
TetR-f CGCTCAAAAGCTGGGAGTTG
TetR-r GCCTGTCCAGCATCTCGATT
GFP-total-f GTGCCCATCCTGGTCGAG
GFP-total-r GTCAGGGTGGTCACGAGG
Microscopic images
HeLa or HeLa HF1–3 cells expressing GFP or A3′SS-T2-CP were grown on glass coverslips for 24 h and transfected with 300 ng TetR-mCherry plasmid using 2 µL of Lipofectamine 2000 (Life Technologies) according to manufacturer instructions. Two hours after transfection, the medium was changed to DMEM with or without 50 µM doxycycline and further incubated at 37°C and 5% CO2 for 24 h. Next, cells were fixed in 3.7% formaldehyde at room temperature (RT) for 10 min followed by permeabilization with 0.5% triton X-100 in 1×PBS for 10 min at RT. Cells were then washed three times with 1× PBS and stained with 4′,6-diamidino-2-phenylindole (DAPI) dye (1 µg/mL in water; Thermo Scientific) and washed three times with 1x PBS. Finally, coverslips were mounted with mounting medium (Thermo Scientific). All experiments were carried out on a Zeiss Axiovert 200 M inverted microscope using a 63×/1.4 NA oil objective lens (Zeiss). Excitation was done using the mercury arc lamp (Zeiss). The following filters were used: 350/50 (excitation) and 460/50 (emission) for DAPI, 482/18 (excitation) and 520/28 (emission) for GFP, and 565/30 (excitation) and 620/60 (emission) for mCherry. Images were repeated three times. Data processing was performed using ImageJ.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Britta Schreiber for technical assistance, Julia Wellstein for her help with fluorescence microscopy, and Leon Kraus for his help with the preparation of Figure 2. This project was funded by Marie Skłodowska-Curie-Grant no. 642738 of the research and innovation program Horizon 2020 of the European Union and the DFG (CRC902 A2).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077453.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Atanasov J, Groher F, Weigand JE, Suess B. 2017. Design and implementation of a synthetic pre-miR switch for controlling miRNA biogenesis in mammals. Nucleic Acids Res 45: e181 10.1093/nar/gkx858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle FE, Giudice J. 2017. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18: 437–451. 10.1038/nrm.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens C, Lochner S, Lober S, Usai I, Schmidt A, Drueppel L, Hillen W, Gmeiner P. 2006. Subtype selective tetracycline agonists and their application for a two-stage regulatory system. ChemBioChem 7: 1320–1324. 10.1002/cbic.200600226 [DOI] [PubMed] [Google Scholar]
- Chen L, Weinmeister R, Kralovicova J, Eperon LP, Vorechovsky I, Hudson AJ, Eperon IC. 2017. Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways. Nucleic Acids Res 45: 2051–2067. 10.1093/nar/gkw860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Reed R. 1999. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402: 207–210. 10.1038/46086 [DOI] [PubMed] [Google Scholar]
- Chua K, Reed R. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol Cell Biol 21: 1509–1514. 10.1128/MCB.21.5.1509-1514.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan SM, Falla A, Goldfless SJ, Nasamu AS, Niles JC. 2016. Synthetic RNA–protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat Commun 7: 10727 10.1038/ncomms10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless SJ, Belmont BJ, De Paz AM, Liu JF, Niles JC. 2012. Direct and specific chemical control of eukaryotic translation with a synthetic RNA-protein interaction. Nucleic Acids Res 40: e64 10.1093/nar/gks028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau FC, Jaeger J, Groher F, Suess B, Muller A. 2020. The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA–TetR regulator complex. Nucleic Acids Res 48: 3366–3378. 10.1093/nar/gkaa083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher AC, Jager S, Schneider C, Groher F, Hamacher K, Suess B. 2019. Tuning the performance of synthetic riboswitches using machine learning. ACS Synth Biol 8: 34–44. 10.1021/acssynbio.8b00207 [DOI] [PubMed] [Google Scholar]
- Gusti V, Kim D, Gaur RK. 2008. Sequestering of the 3′ splice site in a theophylline-responsive riboswitch allows ligand-dependent control of alternative splicing. Oligonucleotides 18: 93–99. 10.1089/oli.2007.0107 [DOI] [PubMed] [Google Scholar]
- Guth S, Martínez C, Gaur RK, Valcárcel J. 1999. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol Cell Biol 19: 8263–8271. 10.1128/MCB.19.12.8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallegger M, Sobala A, Smith CWJ. 2010. Four exons of the serotonin receptor 4 gene are associated with multiple distant branch points. RNA 16: 839–851. 10.1261/rna.2013110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R, Cautain B, de Pedro N, Link W. 2014. Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget 5: 11–28. 10.18632/oncotarget.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS. 2012. The mechanism of the second step of pre-mRNA splicing. Wiley Interdiscip Rev RNA 3: 331–350. 10.1002/wrna.112 [DOI] [PubMed] [Google Scholar]
- Hung M-C, Link W. 2011. Protein localization in disease and therapy. J Cell Sci 124: 3381–3392. 10.1242/jcs.089110 [DOI] [PubMed] [Google Scholar]
- Hunsicker A, Steber M, Mayer G, Meitert J, Klotzsche M, Blind M, Hillen W, Berens C, Suess B. 2009. An RNA aptamer that induces transcription. Chem Biol 16: 173–180. 10.1016/j.chembiol.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Jans DA, Martin AJ, Wagstaff KM. 2019. Inhibitors of nuclear transport. Curr Opin Cell Biol 58: 50–60. 10.1016/j.ceb.2019.01.001 [DOI] [PubMed] [Google Scholar]
- Jenkins JL, Agrawal AA, Gupta A, Green MR, Kielkopf CL. 2013. U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res 41: 3859–3873. 10.1093/nar/gkt046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-S, Gusti V, Pillai SG, Gaur RK. 2005. An artificial riboswitch for controlling pre-mRNA splicing. RNA 11: 1667–1677. 10.1261/rna.2162205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-S, Gusti V, Dery KJ, Gaur RK. 2008. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol Biol 9: 23 10.1186/1471-2199-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Han M-E, Oh S-O. 2017. The molecular mechanism for nuclear transport and its application. Anat Cell Biol 50: 77–85. 10.5115/acb.2017.50.2.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Plass M, Pérez-Valle J, Eyras E, Vilardell J. 2011. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell 43: 1033–1039. 10.1016/j.molcel.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Mol AA, Groher F, Schreiber B, Rühmkorff C, Suess B. 2019. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module. Nucleic Acids Res 47: e132 10.1093/nar/gkz753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, Di Ventura B. 2014. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun 5: 4404 10.1038/ncomms5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B. 2016. Optogenetic control of nuclear protein export. Nat Commun 7: 10624 10.1038/ncomms10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Valle J, Vilardell J. 2012. Intronic features that determine the selection of the 3′ splice site. Wiley Interdiscip Rev RNA 3: 707–717. 10.1002/wrna.1131 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber M, Arora A, Hofmann J, Brutschy B, Suess B. 2011. Mechanistic basis for RNA aptamer-based induction of TetR. Chembiochem 12: 2608–2614. 10.1002/cbic.201100503 [DOI] [PubMed] [Google Scholar]
- Tiebel B, Radzwill N, Aung-Hilbrich LM, Helbl V, Steinhoff HJ, Hillen W. 1999. Domain motions accompanying Tet repressor induction defined by changes of interspin distances at selectively labeled sites. J Mol Biol 290: 229–240. 10.1006/jmbi.1999.2875 [DOI] [PubMed] [Google Scholar]
- Vigevani L, Gohr A, Webb T, Irimia M, Valcárcel J. 2017. Molecular basis of differential 3′ splice site sensitivity to anti-tumor drugs targeting U2 snRNP. Nat Commun 8: 2100 10.1038/s41467-017-02007-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Weigand JE, Kluge B, Grez M, Suess B. 2018. A small, portable RNA device for the control of exon skipping in mammalian cells. Nucleic Acids Res 46: 48–59. 10.1093/nar/gky062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Baxter JE, Wattam SL, Hayward DG, Fardilha M, Knebel A, Ford EM, Da Cruz E, Silva EF, Fry AM. 2007. Alternative splicing controls nuclear translocation of the cell cycle-regulated Nek2 kinase. J Biol Chem 282: 26431–26440. 10.1074/jbc.M704969200 [DOI] [PubMed] [Google Scholar]
- Zillmann M, Zapp ML, Berget SM. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol 8: 814–821. 10.1128/MCB.8.2.814 [DOI] [PMC free article] [PubMed] [Google Scholar]