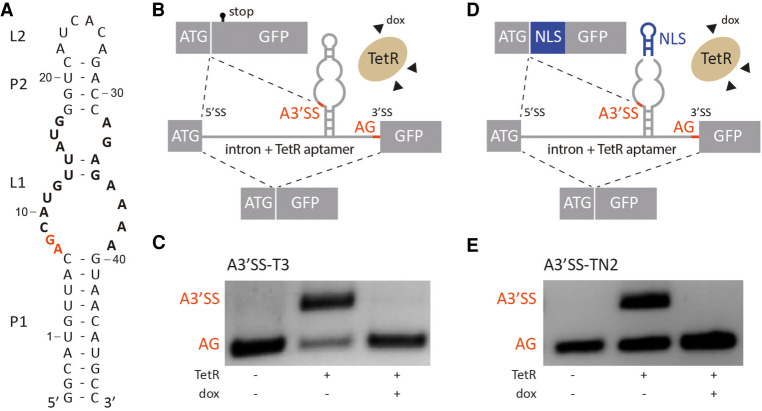
FIGURE 1.
TetR aptamer controls alternative 3′ splice site recognition. (A) Secondary structure of the TetR aptamer. Stems and loops are indicated with P and L, respectively. Nucleotides involved in TetR binding are highlighted in bold, the A3′SS A7G8 is labeled in red. (B,D) Schematic of the proposed model. In the absence of TetR, canonical AG is recognized by the spliceosome. When TetR binds to the aptamer, the A3′SS is activated. The splicing pattern can be restored with the addition of doxycycline (dox) that leads to conformational changes of TetR and the release of the aptamer. Controlled production of two splice variants is possible with this mechanism. Additionally, insertion of a nuclear localization sequence (NLS, in blue) between A3′SS and AG (D) allows to produce different splice variants with different subcellular localization on demand. Exons are displayed as boxes and the intron with the TetR aptamer is shown as a line. (C,E) Splicing pattern visualized by RT-PCR. HeLa cells were transiently transfected with the constructs A3′SS-T3 and A3′SS-TN2 and cotransfected with plasmid expressing TetR (+) and treated with (+) or without (−) 50 µM doxycycline (dox) for 24 h. Total RNA was prepared and used for RT-PCR with primer pairs binding to both exons. The upper band corresponds to usage of A3′SS and the lower band to distal AG. The experiment was repeated three times with similar results.