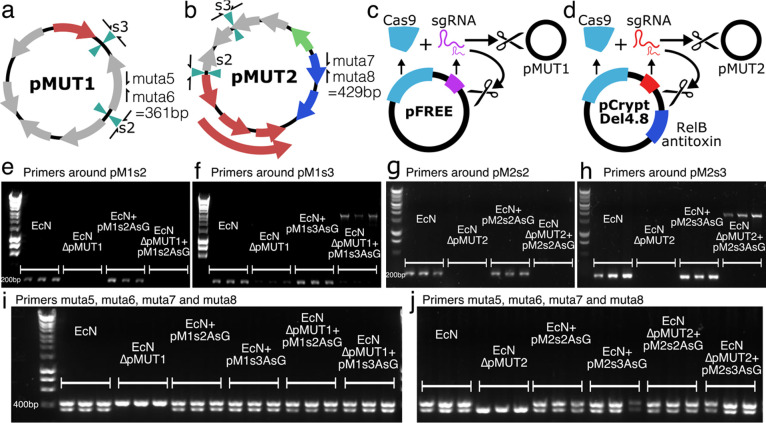
Figure 2.
Curing native pMUT plasmids. EcN pMUT plasmids were assessed with primers around the insertion sites and primers muta5 and muta6 on pMUT1 (a), and muta7 and muta8 on pMUT2 (b). (c) Plasmid pFREE cleaves pMUT1 through expression of Cas9 and gRNA targeting the colE1 origin of replication on pMUT1 and on pFREE itself. (d) Similarly, pCryptDel4.8 targets the origin of pMUT2 and itself, and also contains a RelB antitoxin to disrupt the RelE-RelB toxin-antitoxin system on pMUT2. (e−h) Agarose gels showing the results of colony PCRs around the insertion sites of the “AsG” cassette, revealing that transformation with an engineered plasmid does not displace the native plasmid. (i,j) pFREE and pCryptDel4.8 can cure EcN of native plasmids, and these can be replaced with engineered versions. In all agarose gel images, each condition is shown in triplicate, each lane representing a PCR result using a distinct bacterial colony.