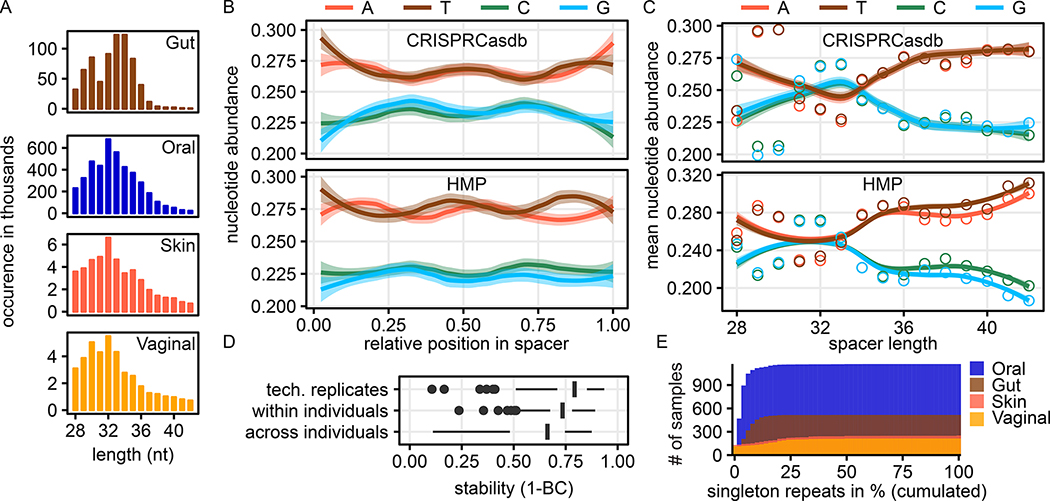
Figure 1: High consistency and agreement of spacer sequences from HMP to public databases and presence of a length-specific GC bias.
A) Sequence lengths of spacers were largely consistent between the minimum of 28 nucleotides and a tail permitted up to 43 nucleotides over different body areas. B) HMP spacers were highly similar to CRISPRCasdb spacers in position-wise nucleotide composition normalised by spacer length and showed a palindromic pattern in both datasets. C) Nucleotide composition stratified by spacer length showed a consistent pattern for HMP- and CRISPRCasdb-derived spacer sequences D) Stability of repeat sequences (as measured by Bray–Curtis dissimilarity of k-mer counts of repeat sequences) across (i) technical replicates, (ii) samples taken from the same individuals over time and (iii) between individuals randomly selected individuals, respectively. Samples containing fewer than 25 repeats are not shown. E) HMP samples generally contain few CRISPR repeats that are sample-specific (singleton repeats). Histogram shows the proportion of singleton repeats among all repeats per sample for all samples.