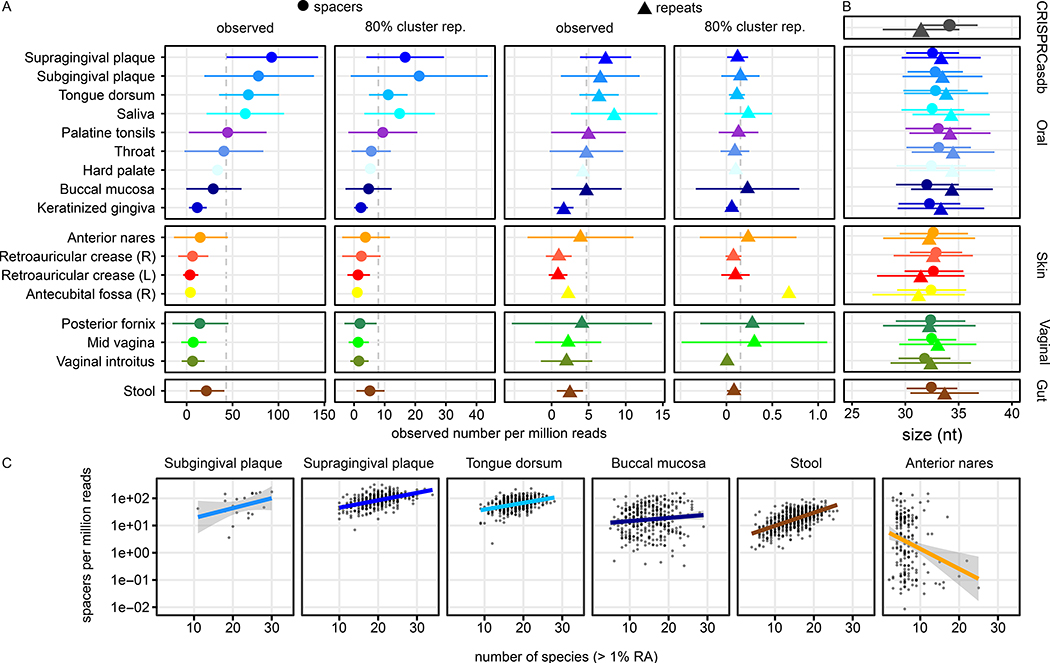
Figure 2: High body-site dependent differences in spacer loads (regardless of host or target) on the HMP1-II dataset.
A) Three oral associated body sites, supra- and subgingival plaque and tongue, have significantly increased CRISPR spacer counts (Wilcoxon rank sum test on spacer counts, P < 10−6) relative to other body sites, such as the urogenital and skin microbiota. Mean values (points for spacers and triangles for repeats) and SD (lines) of the read-depth normalised load per body site are shown for observed reads and repeat and for cluster representatives to account for repetitive sequences. B) The lengths of observed CRISPR spacer and repeat sequences are consistent between most body sites, especially between gut and oral samples, but different from the spacers and repeats present in CRISPRCasdb. Mean (points and triangles) and SD (line) sizes of the spacer and repeat sequences across body sites and within CRISPRCasdb (grey). C) Correlation of species richness (number of species exceeding 1% RA) and spacer load (cluster representatives, defined as the longest sequence within a cluster of > 80% of sequence identity) of selected samples.