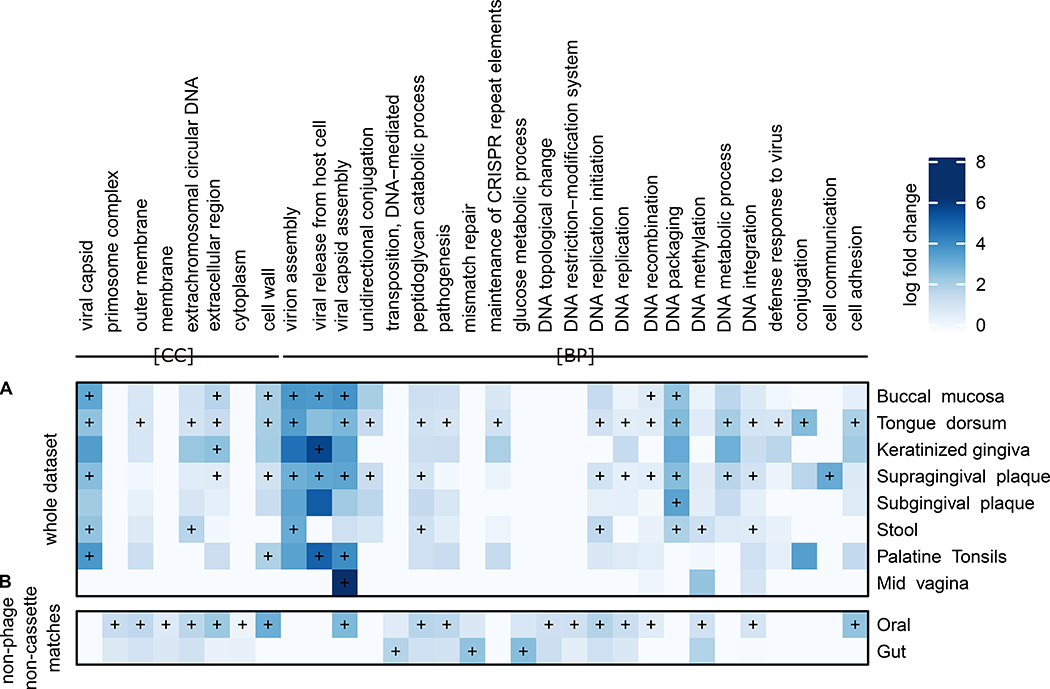
Figure 4: Functional enrichments within predicted spacer targets.
A) Log fold enrichment of Gene Ontology (GO) terms for all spacer targets within sample assemblies per body site. Terms shown here achieved at least one FDR corrected q value < 0.05 based on a Fisher test of enriched UniRef90 terms with respect to the overall contig annotation of the site (STAR Methods). Corresponding spacer targets by the global UniRef90 approach of remaining spacers are shown in Fig. S7) B) GO terms of spacers matching contigs outside of CRISPR-cassettes without any phage-related term on whole contigs, thus potentially within bacterial chromosomes. In both panels, a plus sign denotes an q value < 0.05 and GO groups cellular component (CC) and biological processes (BP) are shown (full version shown in Fig. S8.