Abstract
The coronavirus disease 2019 (COVID-19) pandemic has been a major public health challenge in 2020. Early diagnosis of COVID-19 is the most effective method to control disease spread and prevent further mortality. As such, a high-precision and rapid yet economic assay method is urgently required. Herein, we propose an innovative method to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using isothermal amplification of nucleic acids on a mesh containing multiple microfluidic pores. Hybridization of pathogen DNA and immobilized probes forms a DNA hydrogel by rolling circle amplification and, consequently, blocks the pores to prevent fluid movement, as observed. Following optimization of several factors, including pore size, mesh location, and precision microfluidics, the limit of detection (LOD) for SARS-CoV-2 was determined to be 0.7 aM at 15-min incubation. These results indicate rapid, easy, and effective detection with a moderate-sized LOD of the target pathogen by remote point-of-care testing and without the requirement of any sophisticated device.
Keywords: COVID-19, Rolling circle amplification, Micropores, Nucleic acid, Hydrogel, Microfluidics
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic (Wang et al., 2020a, Wang et al., 2020b) has claimed several lives, particularly among the elderly and those with underlying diseases (Leng et al., 2020; Song et al., 2020; Wang et al., 2020a, Wang et al., 2020b), shattered health care and social systems, and crippled economies on an unprecedented global scale (Kupferschmidt, 2020). Globally, more than 60 million people have been infected, with over one million fatalities in less than a year (WHO, 2020). Due to symptoms similar to those of the common flu, including fever, cough, and sore throat (Hu et al., 2020; Singhal, 2020), most patients with the flu are regarded as COVID-19 suspects and require a molecular-precision virus test, which is highly expensive. Massive and timely testing is also required at airports and public health centers to effectively prevent and control infectious disease (Ding et al., 2020; Gayet et al., 2020; Jahanban-Esfahlan et al., 2019; Pokhrel et al., 2020). Moreover, a highly precise and economic virus diagnostic test is required that can easily be used in developing countries to tackle infectious viral diseases without incurring a financial burden.
Many devices for the diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have been introduced using a range of different amplification and detection methods (Ji et al., 2020; Jin et al., 2020; Vandenberg et al., 2020). To date, reverse transcription polymerase chain reaction (RT-qPCR) (Smyrlaki et al., 2020; Tahamtan and Ardebili, 2020; Xiao et al., 2020) and antibody-based detection (Cady et al., 2020; Vandenberg et al., 2020) have been applied clinically for COVID-19 detection. Antibody-based detection (immunoassay) is limited by its low sensitivity and dependence on a specific antibody. RT-qPCR has the highest sensitivity and accuracy, rendering it the most widely used assay in laboratory environments. The PCR assay requires sample preparation, synthesis of cDNA from viral RNA, a repetitive thermal cyclic process, and highly sensitive detectors, such as fluorescence detectors. Thus, the PCR assay requires specialized facilities, including a laboratory environment, skilled operators, and contamination-free operation. Considering the limitations of this test, it is clear that a cost-effective, and highly accurate method that can serve as an ultra-sensitive point-of-care (POC) device is required to detect COVID-19.
The incorporation of leading-edge technology into a microfluidic platform (Funari et al., 2020; Palestino et al., 2020; Qin et al., 2020) may provide solutions for these urgent issues. The development of several isothermal amplification techniques— specifically loop-mediated isothermal amplification of DNA (LAMP) (Notomi et al., 2000), smart amplification process (Mitani et al., 2007), and rolling circle amplification (RCA)(Huang et al., 2020; Jarvius et al., 2006; Jung et al., 2016; Sato et al., 2010, 2013; Wang et al., 2020a, Wang et al., 2020b)— have been revived due to their enhanced performance, rapid analysis, high sensitivity, and reliable quantification. There have been several recent noteworthy studies on COVID-19 detection using LAMP (Crone et al., 2020; Ganguli et al., 2020; Thi et al., 2020; Thompson and Lei, 2020), including a colorimetric assay and a handheld instrument. However, these detection tests are not fully applicable for virus detection in terms of time and limit of detection (LOD). An RCA assay has demonstrated significant progress with the adoption of a dumbbell-shaped padlock probe, thus resulting in DNA gelation and flow stoppage in a microfluidic system (Goy et al., 2019; Nam et al., 2019; Song et al., 2019). Unfortunately, these assays suffered due to a relatively low LOD (~0.1 pM) and increased test duration (~2 h), indicating significant inferiority to the conventional PCR method, even though the amplification rates of the isothermal assays were greatly enhanced (Ali et al., 2014; Cheng et al., 2009).
Herein, we describe an innovative microfluidic system for fast and ultrasensitive detection of SARS-CoV-2 within 5 or 15 min using DNA concentrations of 3.0 or 30 aM, respectively. The innovative features of the present assay stem from the integration of core techniques, including a continuous binding target during RCA, DNA gelation via RCA, blockage by the DNA gel of the micropores of a thin mesh, and sensitive hydrostatic microfluidics. The test time was innovatively reduced by adopting micro-scale gaps formed in the mesh, which can be easily and rapidly blocked with RCA gelation. Most importantly, due to the simple operation principle, the current system can be operated at any place or POC, even without available electricity.
2. Experimental procedures
The materials used in this study are described in the supplementary information. Throughout this study, all experiments performed 5 replicate tests (n = 5) in each condition.
2.1. Primer immobilization on nylon mesh surface
First, various meshes of different pore sizes (0.2 to 20 μm) and materials were systematically examined in terms of repeatability, test time, and conjugating primers. A nylon mesh with 1-μm pores was finally chosen and used for the present study. Fig. S1 and Fig. S2 describe the selection of an appropriate mesh type and pore size. Prior to immobilization, the nylon mesh was cleaned with 1 mL of isopropyl alcohol and deionized (DI) water. For primer coupling on nylon mesh surface, the nylon mesh was suspended in 380 μL of coupling buffer (0.2 M NaHCO3, 0.5 M NaCl, pH 8.3) and 20 μL of 1 mM NH2 primer. The nylon mesh was then incubated in this solution for 2 h at room temperature (~23 °C). The mesh was then re-incubated in 500 μL of blocking buffer (0.5 M ethanolamine, 0.5 M NaCl, pH 8.3) for 1 h at room temperature (~23 °C). Finally, the primer-conjugated mesh was washed with 1 mL of 1 × phosphate-buffered saline (PBS) solution to remove unbound primer. A detailed description of primer immobilization on the mesh surface is provided in the supplementary information. Also, nucleic acid sequences of template, primer, and pathogen used in this study were listed in Table S1.
2.2. Validation of primer immobilization on nylon mesh surface
A fluorescent probe was used to confirm primer immobilization on the mesh surface. A fluorescent probe solution (5 μL, 1 mM) was mixed with 195 μL of mesh suspension and incubated at 60 °C for 40 min for hybridization. After incubation, the nylon mesh was washed three times with 1 × PBS buffer. The nylon mesh surface was observed by fluorescence microscopy (Olympus 1 × 71, Tokyo, Japan).
2.3. Preparation of templates for immobilization on primer-coupled nylon mesh
Single-stranded DNA template (50 μL, 4 μM) was incubated at 95 °C for 5 min, and then at 60 °C for 3 min to create dumbbell-shaped templates. Suspension primer-immobilized nylon meshes (20 μL) were added to the template solution at 60 °C, which was progressively cooled to 4 °C, using a thermal cycler at 0.5 °C/min. The nylon meshes were washed twice with 200 μL of 1 × PBS.
2.4. Nylon mesh attachment on the microchannel
A nylon mesh was immobilized with primers and then with the padlock probe. The nylon mesh was then punched with its diameter according to the size of the test tube. Then, the circular nylon mesh was assembled with a test tube with a silicon tubing adaptor. The dimensions of the test tube were either 0.5 or 1.0 mm in diameter and 45 mm in length. The nylon mesh-attached tube was then inserted into the microfluidic kit (RSD-K01; Rheomeditech, Inc., Seoul, Korea). Finally, the test tube with the nylon mesh was orientated to the sample chamber, in which the binding probability between the padlock probes and pathogens was maximized with gentle stirring.
2.5. Experimental setup
The microfluidic system to detect SARS-CoV-2 using a mesh-based RCA process is shown in Fig. 1 (a). The system consists of a sample chamber, a glass tube, a padlock probe conjugated nylon mesh, and a waste chamber with a rubber lid. A sample loaded into the sample chamber cannot flow through the test tube because the waste chamber is sealed with a rubber lid, as shown in Fig. 1(b). When the rubber lid is connected to the atmosphere, the sample fluid is driven to the waste chamber due to the elevated hydrostatic pressure in the sample chamber. However, when the membrane pores were blocked with DNA hydrogels, the flow was interrupted, even if the lid was punctured with a syringe needle (Fig. S3). The porous mesh, which was flush-mounted at the entrance of the glass tube (Fig. 1(c)), was directly exposed to the sample during the test. A flexible silicone tube was used for a tight seal in the connection between the test tube and the two chambers; Fig. 1(d) shows the actual image of the nylon mesh mounted on the tube entrance. The micro-holes (~1 μm size) of the nylon mesh were densely but uniformly distributed across the entire mesh surface. These micro-holes were rapidly blocked by DNA entanglement through the RCA process. Fig. 1(e) and (f) show the SEM images of the nylon mesh at two different magnifications.
Fig. 1.
(a) An explanatory schematic of the experimental setup for COVID-19 detection. (b) 3D photograph of the experimental apparatus. (c) 3D photograph of a flexible silicon tube bonded to the nylon mesh and glass tube. (d) Front-view of the attached nylon mesh with silicon tubing to prevent leaks. (e) Scanning electron image of the nylon mesh. (f) Scanning electron image of a single micro-hole of the nylon mesh. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The sample chamber was first filled with a sample fluid containing a colored dye, which provided a visual indication of whether the test tube was blocked or not. To prevent evaporation during a test, a light-density mineral oil was additionally loaded on the sample fluid. When a target pathogen was present in a sample, the pathogen DNA could bind to the padlock probe on the mesh, and a subsequent RCA process occurred. Gentle mixing with a stirrer significantly enhances the binding between the target pathogen and the template. Throughout the RCA process, amplified DNA products become entangled with each other to form DNA hydrogels, which blocked flow paths in the mesh, as shown in Fig. 1(f). After a certain time to allow completion of the RCA process, the rubber lid was punctured with a syringe needle, and the fluid motion and travel time to reach the other end of the test tube were precisely monitored. The travel time (t tr) for a negative control sample that did not contain any pathogens was used as the reference value. A sample containing target pathogens resulted in an extended travel time compared with the reference value. In samples with a high concentration of pathogens, flow paths through the mesh were completely blocked, and thus there was no flow to the end of the test tube (Fig. S3).
2.6. RCA reaction on nylon mesh surface
A schematic diagram of the formation of DNA hydrogel by RCA on the nylon mesh surface for SARS-CoV-2 detection is shown in Fig. 2 . Most of the processes are similar to our previous concept of forming DNA hydrogel via RCA, except for the presence of a mesh (Na et al., 2018). The unique difference involves the use of a very thin porous mesh instead of a thick porous hydrogel layer. A brief description of the proposed scheme is as follows: First, a primer was conjugated with the nylon mesh via surface activation and functionalization (Fig. 2(a)). Then, a padlock probe was hybridized with the primer (Fig. 2(b)). The padlock probe was carefully designed to include a primer-binding site, a pathogen-binding site, and a self-assembly region that forms a molecular dumbbell shape. The padlock probe was constructed with consideration for thermal stability at room temperature (~23 °C) and specificity for primer and pathogen hybridization by controlling the length of each binding region. Upon annealing, the padlock probe formed an asymmetric dumbbell shape. The probe was then hybridized with the primer immobilized on the nylon mesh surface. When the padlock probe encountered a target pathogen, hybridization occurred. Using a ligase, the opened padlock probe was ligated to form a closed-loop template, which could then undergo the RCA process. As time passed, complementary single-stranded DNA were elongated with a dumbbell shape by Bst 3.0 DNA polymerase during the RCA process. Due to the dumbbell-shaped template, amplified long DNAs tended to easily entangle with one another, aggregating with neighboring DNAs and forming a DNA gel, as shown in Fig. 2 (f). Further elaboration is given in the supplementary information (Fig. S4).
Fig. 2.
Schematic of DNA hydrogel formation through rolling circle amplification using nylon mesh. (a) Templates are self-assembled to form an asymmetric dumbbell shape, and primers are immobilized on the nylon mesh surface. (b) Templates are hybridized with primers immobilized on nylon mesh surface. (c) When the template on the nylon-mesh surface hybridizes with a target pathogen, (d) the template is ligated to form a closed-loop template. (e) DNA hydrogel formation by COVID-19 template DNA (g) RCA products are elongated by Phi29 polymerase. (f) Polyacrylamide gel electrophoresis analysis of COVID-19 template structures. M, markers. 1) pathogen, 2) linear template, 3) self-assembled dumbbell-shape template, 4) template hybridized with a target pathogen, 5) closed-loop template formed by ligation, and 6) amplified RCA products.
All intermediate products from the stepwise processes, including template structure formation, primer-probe hybridization, ligation, and amplification using polyacrylamide gel electrophoresis (PAGE), are shown in Fig. 2(g). 1: A pathogen DNA with a length of 20 nucleotides (nt) was observed at the bottom of the gel. 2: A relatively long single-stranded padlock-probe DNA (102 nt) was observed near the middle of the gel. 3: When the padlock probe self-assembled by annealing to form an asymmetric dumbbell shape, a band was observed above the linear template band due to the complexity of the structure. 4: When the pathogen DNA hybridized to the dumbbell-shaped probe, the double-stranded DNA portion increased, and a band was observed at a higher position. 5: When the padlock probe formed the closed loop structure through the ligation process, a band was observed at a higher position than that before ligation. 6: Final amplified RCA product; further detailed protocol is provided in the supplementary information. The effect of incubation time on the RCA process is also elaborated in the supplementary information (Fig. S5), whereas the reasons for selection of Bst 3.0 DNA polymerase as well as T7 ligase are discussed in Fig. S6.
2.7. Rheological measurements of RCA products
We used a microfluidic rheometry, Rheoscan-D300 (Rheomeditech, Inc., Seoul, Korea), which can measure viscosity over a range of shear rates (0.1–1000 s−1) as well as yield shear stress. Using the same protocol for gel formation, we prepared gels at incubation times of 0, 30, 60, and 120 min at a fixed pathogen concentration of 3 fM.
3. Results and discussion
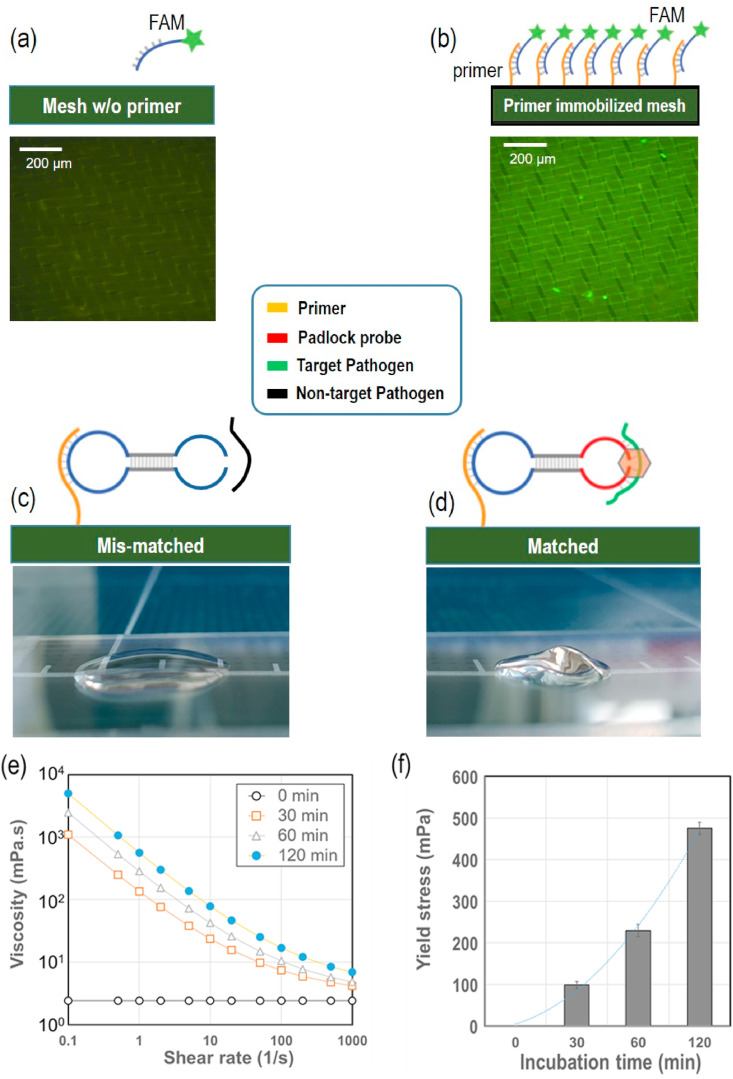
The present study adopted multi-step precision processes, each of which required validation. First, we examined the primer immobilization effect via fluorescence microscopy (Olympus 1 × 71, Tokyo, Japan). For this, the nylon mesh with or without primer immobilization was compared, as shown in Fig. 3 (a) and (b). Successful immobilization of primers on the nylon mesh surface enabled combination with the complementary fluorescent probe (FAM). The nylon mesh immobilized with the primer produced bright fluorescent images, whereas the nylon mesh without the immobilized primer did not. These results clearly confirm that the nylon mesh was precisely immobilized with the desired primer.
Fig. 3.
Validation of DNA gelation on nylon mesh surface. (a) Schematic of nylon mesh surface without primer immobilization and hybridization with the fluorescence probe FAM; a 200-μm fluorescence microscopic image is also shown (lower image). (b) Schematic of nylon mesh surface with primer immobilization and hybridization with the fluorescence probe FAM; a 200 μm fluorescence microscopic image is also shown (lower image). (c) Mobile phone-captured image of a mismatched experiment; Non-target pathogen (dengue pathogen) was used with the COVID-19 DNA template. (d) Mobile phone-captured image of a matched experiment; target pathogen (COVID-19 pathogen) was used with the COVID-19 DNA template. (e) Increase in viscosity with incubation time (0, 30, 60, and 120 min) (n = 5), (f) Increase in yield shear stress with incubation time (0, 30, 60, and 120 min).
To confirm the specificity of the pathogen detection, two different virus templates (SARS-CoV-2 and dengue virus) were examined with the present padlock probe. Detection of other viruses such as Ebola and MERS has also been assessed using the present padlock probes without hydrogel formation due to mismatches between targets and probes (Na et al., 2018). The padlock probes were not homologous with the dengue virus DNA sequence and did not form a hydrogel, as shown in Fig. 3(c). When the present padlock probes encountered SARS-CoV-2 DNA, a strong RCA reaction occurred, followed by apparent hydrogel formation, as shown in Fig. 3(d).
To quantify the extent of difference in gelation, we characterized the rheological properties of the solutions with the COVID-19 pathogen target. As shown in Fig. 3(e), the solution viscosity significantly increased with incubation time, showing a strong shear-thinning characteristic. At a low shear rate of 0.1 s−1, the viscosity at 60-min incubation increased more than 1000 folds compared with that at 0 min. It is important to note that yield shear stress was significantly generated with gel formation, as shown in Fig. 3(f). This significantly increased yield stress caused by DNA gelation was sufficient to resist driving pressure and prevent flow through the membrane micropores. The yield stress of fluids is frequently encountered in various fluids, including toothpastes, blood, foams, mud, and polymeric liquids. The common feature of these fluids is that they are able to flow only if they are subjected to a stress above some critical value (i.e., yield stress). Otherwise, under shear stress below the yield stress, they behave in a solid deformation manner. As the applied pressure on the nylon mesh was approximately 97 Pa (10 mmH2O), which was below the yield stresses measured in Fig. 3(f), no flow resulted, which is in accord with the yield stress analysis.
The sensitivity of the present assay was carefully examined in terms of limit of detection (LOD) with varying concentrations of target pathogen templates and incubation times. As described earlier, the measured parameters were the travel time between the inlet and outlet of a test tube and the corresponding velocity. If the mesh pores were completely blocked with amplified DNA gels, the travel time would be infinity and the corresponding velocity would be zero. When the pathogen concentrations were gradually decreased from 300 to 30 zM, the travel time decreased, whereas the corresponding velocity increased (Fig. 4 (a) and (b)). At pathogen template concentrations of 3 nM to 3 fM, there was no flow (infinite travel time or zero velocity), which implies that the mesh pores were completely blocked with DNA gels via the RCA process. Using concentrations of 300 aM to 3 aM, there was flow, but the travel times and velocities significantly differed from those of the reference. However, at 30 and 300 zM template concentrations, the travel times and velocities were nearly the same as those of the reference.
Fig. 4.
Analysis of limit of detection (LOD) with varying concentrations of target pathogen templates and incubation times (n = 5) (a) Travel time with respect to COVID-19 pathogen DNA concentrations of 0 and 31.3 zM to 313 pM. (b) Flow velocity with respect to COVID-19 pathogen DNA concentrations of 0 and 31.3 zM to 313 pM. (c) Incubation time of 0, 5, 15, or 30 min with respect to travel time. (d) Incubation time of 0, 5, 15, or 30 min with respect to ink flow velocity. The travel time for complete blockage of the ink flow is assumed as 300 s. (e) Heat-map for COVID-19 detection in terms of pathogen concentration and incubation time. (*: p < 0.05, **: p < 0.001).
Considering these results, the LOD was determined using the following procedure: Notably, the limit of blank (LoB) is calculated by the meanblank + 1.645 (SDblank), while the LOD is calculated by the LOB + 1.645 (SDlow concentration sample) (Armbruster and Pry 2008). In the present study, the blank was used as the negative control. At a fixed incubation time of 15 min, the negative control (blank) travel time and velocity were 17.2 ± 1.5 s and 5.9 ± 0.5 mm/s, respectively. Thus, the LOB of the travel time and velocity were 19.6 s and 4.7 mm/s, respectively. The travel time and velocity SD at a low concentration (3 aM) were 11.2 s and 0.35 mm/s, respectively; thus, the determined LOD of travel time and velocity were 38.8 s and 4.3 mm/s, respectively. With these values, the LOD of the minimum pathogen concentration was found to be 0.7 aM for the travel time and 0.6 aM for the velocity. These LOD values are valid for a fixed incubation time of 15 min and are more sensitive with increasing incubation time and vice versa. We then examined the minimum test time needed for pathogen detection through the RCA process with varying incubation times. The incubation times were 5, 15, or 30 min for the two pathogen concentrations (3.13 and 31.3 aM). There were significant differences between the travel time and velocity at 5 min and 31.3 aM and 15 min and 3.13 aM compared with the corresponding values of the negative control (p < 0.05), as shown in Fig. 4 (c, d). Thus, both incubation time and concentration should be considered to determine the LOD in the present system, as shown in Fig. 4(e), which depicts the pathogen detection capability with colorimetric indication. All tests were performed more than five times throughout the experiments.
Compared with our previous results (LOD = 0.1 pM) (Na et al., 2018), the present LOD was nearly 106-fold enhanced using the more elaborate design of the microfluidic system. First, the membrane was in contact with the sample chamber, maximizing the opportunity to react with the pathogen throughout the test time. In the previous system, a once-through flow system was adopted; as the probes were located in the middle of the microfluidic channel, gentle mixing was performed with a stirrer to further enhance the binding efficiency. Second, the pore size was optimally minimized for rapid blocking by the RCA-induced hydrogel. The smaller the pore, the quicker the blockage can occur. Several different nano-pore membranes were examined but failed to ensure a repeatable travel time, even for a negative sample, which might be due to non-uniform pore arrangements as well as surface tension. The current membrane is a nylon mesh with 1-μm pores. Third, the driving force of the sample fluid was minimized to avoid breaking the DNA hydrogel-induced sealing on the mesh pores. The previous driving force was 3.65 kPa (373 mmH2O), whereas the present one was 97 Pa (10 mmH2O). Due to this minimized driving force, the sealing test with the DNA hydrogel became more sensitive, and thus, the corresponding LOD was significantly enhanced.
The ongoing COVID-19 pandemic desperately requires a rapid test with high sensitivity. The LOD of a virus assay is an important parameter to determine the sensitivity of a sensor to detect viruses. Due to the emergency situation of the COVID-19 pandemic, the US Food and Drug Administration (FDA) has allowed unapproved in vitro diagnostic devices (IVDs) or unapproved uses of approved IVDs under emergency use authorization (EUA). According to the FDA EUA guidelines, the LOD of IVDs for SARS-CoV-2 is 100,000 copies/mL (or 200 copies/reaction), which is approximately 300 aM (FDA 2020). The LOD value of the present system (0.7 aM) is similar to that of FDA-approved PCR assays. Additionally, testing time should be short enough to manage high numbers of immediate tests at public places, such as airport and border checkpoints. Most FDA-approved devices that adopt thermal cyclic PCR assays require more than an hour for test completion and, thus, cannot be used as rapid POC tests. The test time of the present system at 30 aM and higher concentrations was 5 min, which is sufficiently short to be applicable towards massive testing at POCs.
An essential feature of the present study is the formation of a DNA hydrogel through RCA using a dumbbell-shaped probe template. Due to the dumbbell-shaped template, amplified DNA strands become easily entangled with each other and rapidly form a gel. These DNA hydrogels efficiently block the micropores of the nylon mesh. Unlike other meshes, the current one described herein is composed of a highly dense structure with a minimum opening rate of the 3D structured micropores and, thus, has a relatively small flow resistance, yet regardless is easily blocked with the RCA-induced DNA hydrogel. When a small-diameter glass tube is used, the test repeatability diminishes due to capillary phenomena that are sensitive to material types and surface conditions. Thus, the present system used a glass tube with a diameter of 1 mm.
In addition, we compared gel formation between DNA and RNA pathogens using RCA. Both DNA and RNA pathogens yielded significantly increased extensional properties (i.e., elongation viscosity) via a simple pipetting test (Fig. S7). Furthermore, both showed nearly equivalent performance in terms of travel time and velocity at a fixed concentration of 300 aM for various incubation times (5, 15, or 30 min). The results with a detailed description are given in the supplementary information. We then examined the selectivity of virus detection under fixed optimal conditions, including incubation time, nylon mesh-attached tube length, and amount of ink. The results for the detection of SARS-CoV-2, Ebola virus, Zika virus, dengue virus, and MERS CoV are explained in Table S2 and Fig. S9 (Supplementary Information). The microfluidic chip was equipped with a simple single channel containing a single DNA template of SARS-CoV-2. Incubating the COVID-19 pathogen for 30 min enabled the DNA template to generate a strong DNA hydrogel that completely blocked flow, whereas testing of other pathogens for the same specific incubation time failed to form hydrogels, and the ink flowed freely without any blockage. These results show that the current microfluidic platform can selectively detect specific pathogens without any cross-reaction to DNA from other pathogens.
4. Conclusion
In summary, a rapid and ultrasensitive microfluidic-based biosensor was designed and characterized for specific detection of COVID-19 at POCs. The RCA technique with a nylon mesh medium was efficiently maximized to detect COVID-19. On the surface of the nylon mesh attached to the glass tube, DNA hydrogels are massively formed by RCA and quickly and easily block the flow path in the mesh-attached tube. The key advantage of this technology is the minimum effective surface area between the pathogen target and the source (nylon mesh) and continuous rotation of the magnet bar to attach the maximum number of pathogen molecules. In addition, because the flow paths through the nylon mesh were dramatically decreased with the high density of the micro-hole structures of the mesh, the amount of DNA hydrogel required to block the glass tube or channel reduced. Therefore, the present microfluidic system can detect SARS-CoV-2 with an excellent LOD (~3 aM in 15 min or 30 aM in 5 min), which is the lowest LOD until now. Furthermore, because the present method utilizes RCA as the molecular diagnostic tool, the present system is highly selective and accurate. Practical implementation of the present microfluidic system can be easily applied for screening tests at airports and other locations where infectious diseases commonly spread. Simultaneous multiplex detection of various infectious viruses, specifically those responsible for COVID-19, influenza-A, and influenza-B, using a microfluidic platform is the focus of ongoing research. As a limitation of the present study, the tested samples were not clinical samples obtained from various biofluids such as saliva, mucus, and nasopharyngeal swabs but synthetic templates of nucleic acid. With IRB approval and biosafety facility, a clinical evaluation must be performed for the full credit of the proposed method.
Funding
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korean Government Ministry of Science and ICT (MSIT, Korea) [grant number 2016R1A5A1010148] as well as by the Korea Evaluation Institute of Industrial Technology (KEIT) funded by the Korean Government Ministry of Trade, Industry & Energy (MOTIE, Korea) [grant number 20012427].
CRediT authorship contribution statement
Hwang-soo Kim: Investigation, and, Formal analysis. Naseem Abbas: Investigation, original draft, Methodology, review, and editing. Sehyun Shin: Conceptualization, Supervision, Methodology, Visualization, review, and editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D.-K., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43(10):3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008;29(Suppl. 1):S49. [PMC free article] [PubMed] [Google Scholar]
- Cady N.C., Tokranova N., Minor A., Nikvand N., Strle K., Lee W.T., Page W., Guignon E., Pilar A., Gibson G. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2020;171:112679. doi: 10.1016/j.bios.2020.112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zhang X., Li Z., Jiao X., Wang Y., Zhang Y. Highly sensitive determination of microRNA using target‐primed and branched rolling‐circle amplification. Chem. Int. Ed. Engl. 2009;121(18):3318–3322. doi: 10.1002/anie.200805665. [DOI] [PubMed] [Google Scholar]
- Crone M.A., Priestman M., Ciechonska M., Jensen K., Sharp D.J., Anand A., Randell P., Storch M., Freemont P. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-18130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA U. FDA; 2020. Policy for Coronavirus Disease-2019 Tests during the Public Health Emergency (Revised) [Google Scholar]
- Funari R., Chu K.-Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A., Mostafa A., Berger J., Aydin M., Sun F., Valera E., Cunningham B.T., King W., Bashir R. bioRxiv; 2020. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayet R.V., de Puig H., English M.A., Soenksen L.R., Nguyen P.Q., Mao A.S., Angenent-Mari N.M., Collins J. Creating CRISPR-responsive smart materials for diagnostics and programmable cargo release. Nat. Protoc. 2020;15(9):3030–3063. doi: 10.1038/s41596-020-0367-8. [DOI] [PubMed] [Google Scholar]
- Goy C.B., Chaile R.E., Madrid R. Microfluidics and hydrogel: a powerful combination. React. Funct. Polym. 2019;145:104314. [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020:1–14. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., He L., Li S., Liu H., Jin L., Chen Z., Zhao Y., Li Z., Deng Y., He N. A simple fluorescence aptasensor for gastric cancer exosome detection based on branched rolling circle amplification. Nanoscale. 2020;12(4):2445–2451. doi: 10.1039/c9nr08747h. [DOI] [PubMed] [Google Scholar]
- Jahanban-Esfahlan R., Seidi K., Jahanban-Esfahlan A., Jaymand M., Alizadeh E., Majdi H., Najjar R., Javaheri T., Zare P. Static DNA nanostructures for cancer theranostics: recent progress in design and applications. Nanotechnol. Sci. Appl. 2019;12:25. doi: 10.2147/NSA.S227193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvius J., Melin J., Göransson J., Stenberg J., Fredriksson S., Gonzalez-Rey C., Bertilsson S., Nilsson M. Digital quantification using amplified single-molecule detection. Nat. Methods. 2006;3(9):725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- Ji M., Xia Y., Loo J.F.-C., Li L., Ho H.-P., He J., Gu D. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020;10(56):34088–34098. doi: 10.1039/d0ra04507a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., Deng L., Zheng C., Zhou J., Shi H. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I.Y., You J.B., Choi B.R., Kim J.S., Lee H.K., Jang B., Jeong H.S., Lee K., Im S.G., Lee H. A highly sensitive molecular detection platform for robust and facile diagnosis of Middle East respiratory syndrome (MERS) corona virus. Adv. Healthc. Mater. 2016;5(17):2168–2173. doi: 10.1002/adhm.201600334. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. The lockdowns worked—but what comes next? Science. 2020;368:218–219. doi: 10.1126/science.368.6488.218. [DOI] [PubMed] [Google Scholar]
- Leng L., Cao R., Ma J., Mou D., Zhu Y., Li W., Lv L., Gao D., Zhang S., Gong F. Pathological features of COVID-19-associated lung injury: a preliminary proteomics report based on clinical samples. Signal. Transduct. Target. Ther. 2020;5(1):1–9. doi: 10.1038/s41392-020-00355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y., Lezhava A., Kawai Y., Kikuchi T., Oguchi-Katayama A., Kogo Y., Itoh M., Miyagi T., Takakura H., Hoshi K. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat. Methods. 2007;4(3):257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- Na W., Nam D., Lee H., Shin S. Rapid molecular diagnosis of infectious viruses in microfluidics using DNA hydrogel formation. Biosens. Bioelectron. 2018;108:9–13. doi: 10.1016/j.bios.2018.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Jang W.S., Kim J., Lee H., Lim S. Lamb wave-based molecular diagnosis using DNA hydrogel formation by rolling circle amplification (RCA) process. Biosens. Bioelectron. 2019;142:111496. doi: 10.1016/j.bios.2019.111496. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12) doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palestino G., García-Silva I., González-Ortega O., Rosales-Mendoza S. Can nanotechnology help in the fight against COVID-19? Expert Rev. Anti Infect. Ther. 2020:1–16. doi: 10.1080/14787210.2020.1776115. [DOI] [PubMed] [Google Scholar]
- Pokhrel P., Hu C., Mao H. Detecting the coronavirus (COVID-19) ACS Sens. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., Peng R., Baravik I.K., Liu X. Fighting COVID-19: integrated micro-and nanosystems for viral infection diagnostics. Matter. 2020;3:628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Ishii R., Sasaki N., Sato K., Nilsson M. Bead-based padlock rolling circle amplification for single DNA molecule counting. Anal. Biochem. 2013;437(1):43–45. doi: 10.1016/j.ab.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Sato K., Tachihara A., Renberg B., Mawatari K., Sato K., Tanaka Y., Jarvius J., Nilsson M., Kitamori C. Microbead-based rolling circle amplification in a microchip for sensitive DNA detection. Lab Chip. 2010;10(10):1262–1266. doi: 10.1039/b927460j. [DOI] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Vondracek M., Papanicoloau N., Lentini A., Aarum J., Muradrasoli S., Albert J., Högberg B., Reinius B. medRxiv; 2020. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.-Y., Xu J., He Q., Lu Q. Immune dysfunction following COVID-19, especially in severe patients. Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-72718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Zhang Y., Cheng P., Chen X., Luo Y., Xu W. A rapidly self-assembling soft-brush DNA hydrogel based on RCA products. Chem. Commun. 2019;55(37):5375–5378. doi: 10.1039/c9cc01022j. [DOI] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12(556) doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D., Lei Y. Mini Review: recent progress in RT-LAMP enabled COVID-19 detection. Sens. Actuator. Rep. 2020;2:100017. doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2020;1–13 doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.Q., Kaelber D.C., Xu R., Volkow N.D. COVID-19 risk and outcomes in patients with substance use disorders: analyses from electronic health records in the United States. Mol. Psychiatr. 2020;1–10 doi: 10.1038/s41380-020-00880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease ( COVID-19): Situation Report, 182. [Google Scholar]
- Xiao A.T., Tong Y.X., Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa460. Ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.