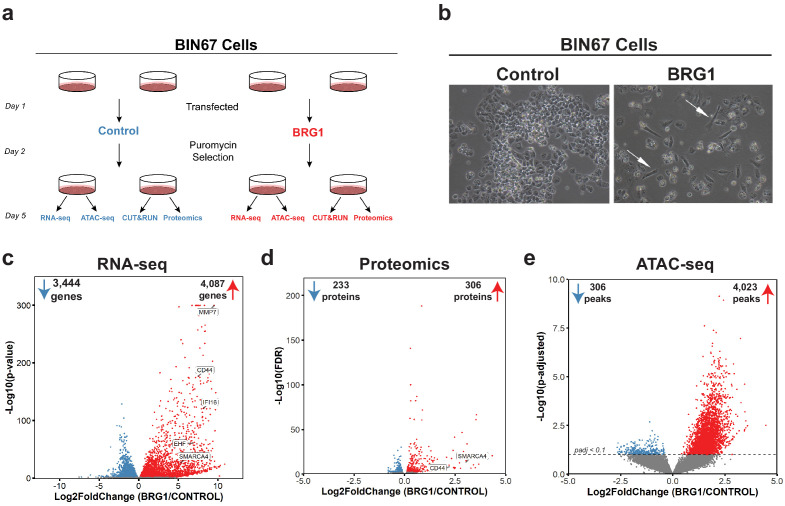
Figure 1. BRG1 reexpression induces global morphological, transcriptomic, and epigenetic changes in BIN67 cells.
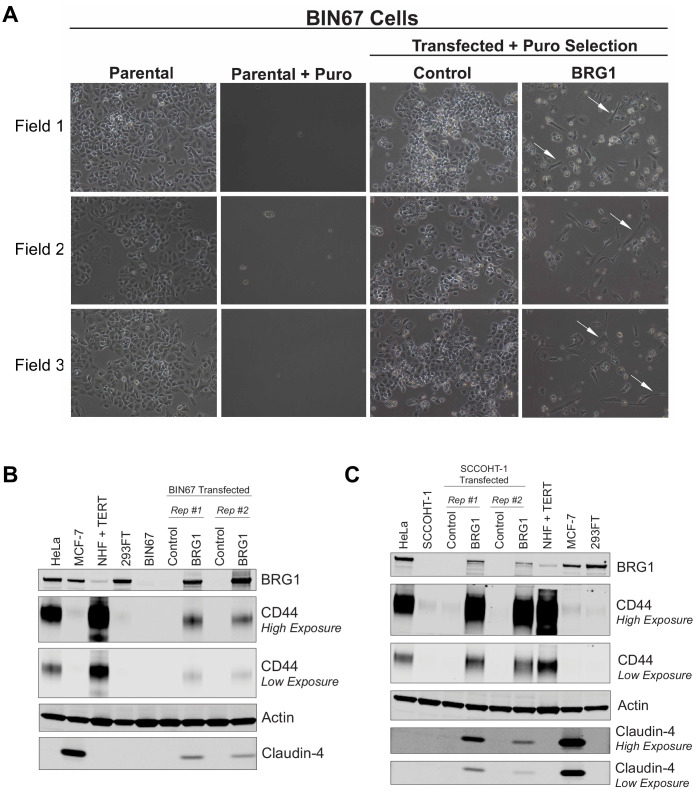
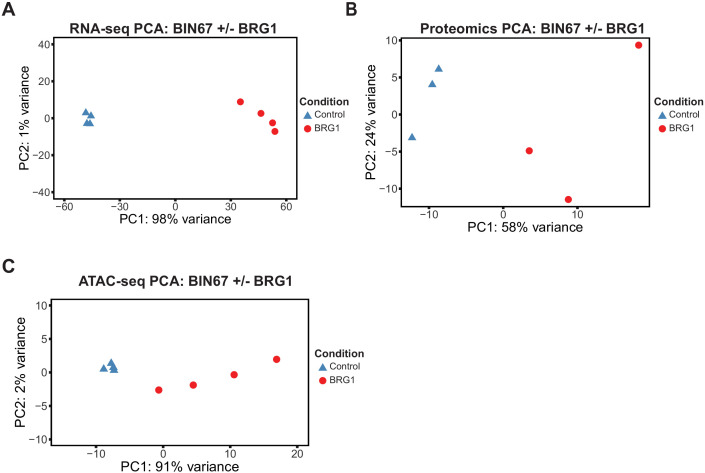
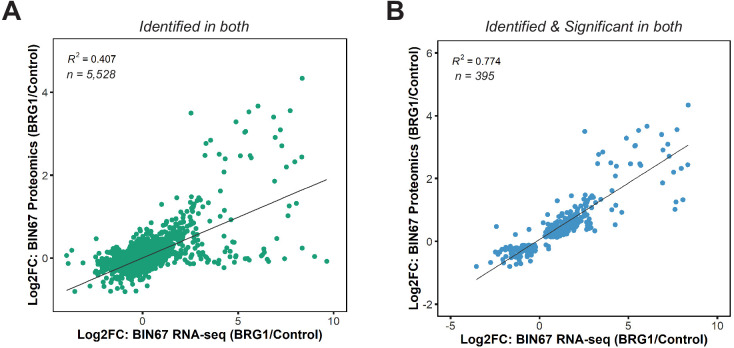
(a) Experimental design for BRG1 reexpression in BIN67 cells and multi-omic data analysis. (b) Following the experimental design in 1a, pictures were taken at 10X phase. BIN67 cells transfected with BRG1 show an elongated morphology (white arrows) relative to control. Additional fields and controls shown in Figure 1—figure supplement 1. (c) Volcano plot results of RNA-seq differential gene expression (BRG1/Control) for protein coding genes using DESeq2 (n = 18,507 genes total). Significantly upregulated genes (padj <0.05 and Log2FoldChange > 0) are colored in red (n = 4087 genes). Significantly downregulated genes (padj <0.05 and Log2FoldChange < 0) are colored in blue (n = 3444 genes). Non-significant genes are colored in gray (n = 10,976 genes). SMARCA4/BRG1 and BRG1 target genes are identified. (d) Volcano plot of differential expressed proteins for BIN67 +/- BRG1 reexpression by PECA analysis (n = 5726 total proteins identified). Significantly upregulated proteins (p.fdr <0.05 and Log2FoldChange > 0) are colored in red (n = 306 proteins). Significantly downregulated proteins (p.fdr <0.05 and Log2FoldChange < 0) are colored in blue (n = 233 proteins). Non-significant proteins are identified in gray (n = 5187 proteins). Proteins of SMARCA4/BRG1 and CD44 are identified. (e) Differential peak analysis for ATAC-seq data using DESeq2 (n = 62,308 peaks total). Significantly gained ATAC-seq peaks (padj <0.10 and Log2FoldChange > 0) are identified in red (Gained; n = 4,023). Significantly lost ATAC-seq peaks (padj <0.10 and Log2FoldChange < 0) are identified in blue (Lost; n = 306). Non-significant peaks are identified in Gray (Static/N.S.; n = 57,979). Source data is available in Figure 1—source data 1.