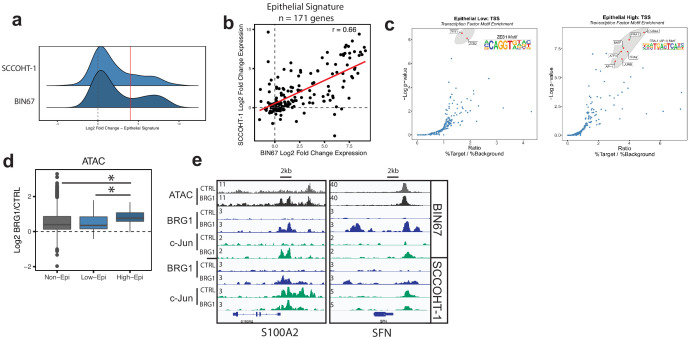
Figure 5. Epithelial genes dependent on AP-1 are highly upregulated by BRG1 expression.
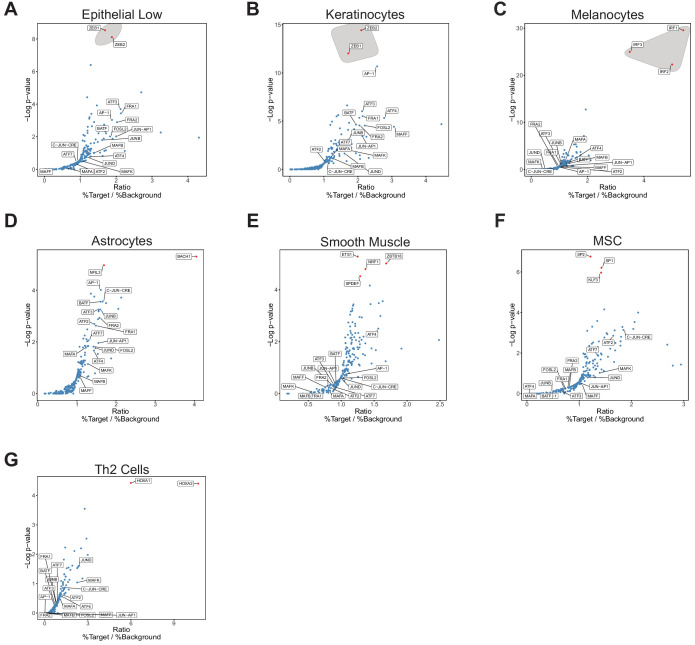
(a) Density plot of RNA-seq Log2FoldChange (BRG1/Control) for epithelial signature genes (n = 171 genes). Red dotted line indicates cutoff point for ‘High’ category (Log2FoldChange > 4). (b) Scatterplot of log2FoldChanges (+/- BRG1) in BIN67 (x-axis) and SCCOHT-1(y-axis). Pearson’s correlation. = 0.66. (c) Known transcription factor motif analysis at the promoter of (b) low epithelial gene and (c) high epithelial genes from BIN67 xCell gene set. Default settings defined by HOMER were used for promoter region (−300 bp, +50 bp from Transcription Start Site, TSS) and background. Transcription factors that were also expressed in RNA-seq data were plotted. Transcription factors that were not expressed at a mean count across all samples greater than four TPMs were eliminated. Highest ranking based on -log(p-value) and Ratio (% Target / % Background) and of similar TF families plotted in red and encircled in gray. (d) Box plot of ATAC-seq signal (Log2FoldChange; BRG1/Control) at the BRG1 CUT and RUN peaks annotated to all high epithelial genes (n = 48 peaks), all low epithelial genes (n = 94 peaks) and non-epithelial genes (n = 15,562 peaks). Non-epithelial genes defined as a BRG1 CUT and RUN-seq peak annotated to nearest gene not contained in either high or low group. Statistical analysis was performed by Wilcoxon Rank Sum Test on low epithelial versus non-epithelial (N.S., p-value=0.907), low epithelial versus high epithelial (p-value=3.5e−4), and high epithelial versus non-epithelial (p-value=5.4e−5). (e) IGV browser tracks of two epithelial signature loci. Source data is available in Figure 5—source data 1.