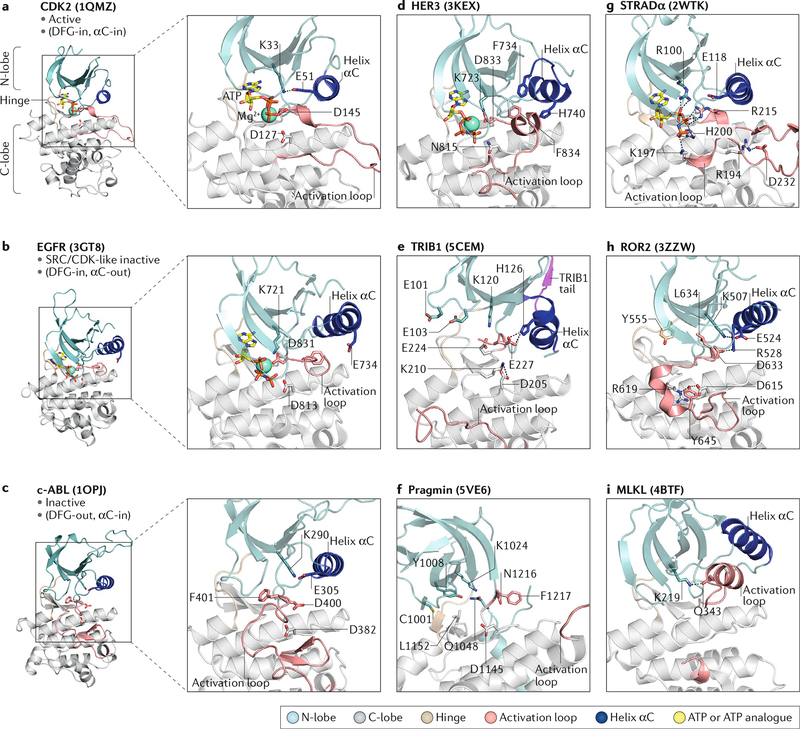
Fig. 3: Structural features of pseudokinases.
Left panels show crystal structures of the CDK2, epidermal growth factor receptor (EGFR) and ABL kinase domains in the active, SRC/CDK-like inactive and inactive conformations, respectively. Insets show zoomed views of the active sites. Right panels show zoomed views of the pseudoactive sites in the crystal structures of the indicated pseudokinases. a | In the active conformation (CDK2 is shown), the β3 lysine (K33) forms a salt bridge with E51 in helix αC, and the DFG (Asp-Phe-Gly) aspartate (D145) coordinates Mg2+ in the active site. b | In the SRC/CDK-like inactive conformation (EGFR is shown), helix αC is shifted away from the active site, preventing a salt bridge between the β3 lysine (K721) and E734. c | In the inactive conformation (ABL is shown), the DFG motif is flipped so that phenylalanine (F401) blocks nucleotide binding. d | Helix αC in human epidermal growth factor receptor 3 (HER3) is partially unwound and adopts a SRC/CDK-like inactive conformation. e | Helix αC is unusually short in TRIB1 and adopts a kink, resulting in a shallow nucleotide-binding pocket. f | Pragmin has a disordered helix αC and occluded nucleotide-binding pocket. g | STE20-related adaptor-α (STRADα) coordinates ATP using positively charged residues. h | The ROR2 DLG motif is shifted ~4 Å compared with canonical DFG motifs and interacts with helix αC, pushing it away from the pseudoactive site. i | Mixed lineage kinase domain-like protein (MLKL) has an unusual activation loop helix that interacts with the β3 lysine (K219). The corresponding Protein Data Bank codes for each structure shown are indicated in parentheses. C-lobe, carboxyl-terminal lobe; N-lobe, amino-terminal lobe.