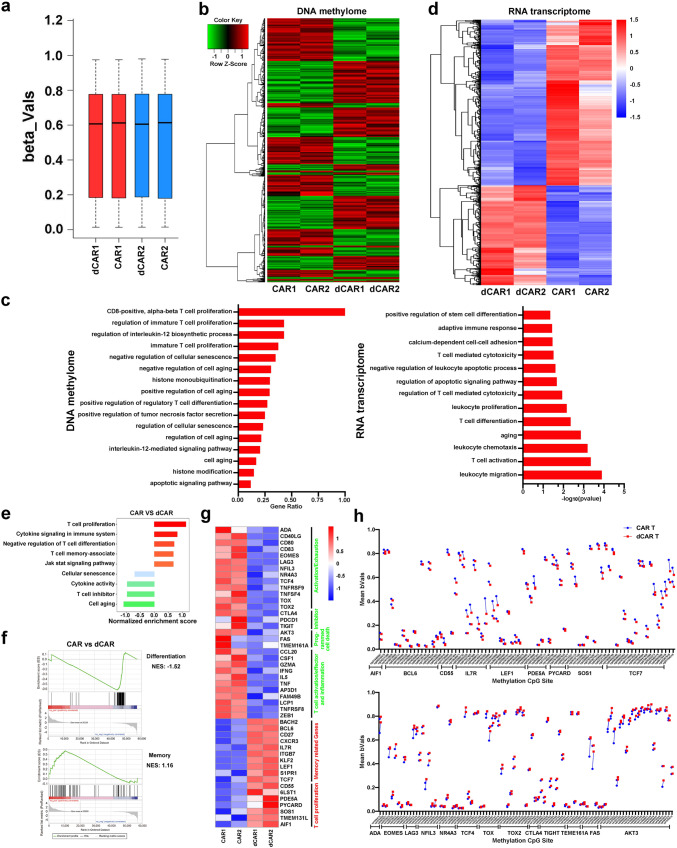
Fig. 2. Transcriptional signatures and DNA methylation characteristics of CAR T cells after DAC treatment.
Gene expression and DNA methylome profiles of CAR and dCAR T cells after 7 days of CAR gene infection. a The methylation status of all the probes was denoted as beta (β) value, which is the ratio of the methylated probe intensity to the overall probe intensity. The mean beta values of CAR T and dCAR T cells measured by chip-based 850k whole-DNA methylome analysis (n = 2) are shown. b Hierarchical clustering analysis of significant differential methylation CpG sites among dCAR T cells compared to CAR T cells (n = 2). c Left: GO functional clustering of significantly differentially expressed CpG site-related genes were enriched in biological processes in dCAR T cells; data are presented as the ratio of the number of CpG-related genes to the number of GO item genes (n = 2). Right: GO functional clustering of genes that were upregulated for biological processes in dCAR T cells (n = 2). d Hierarchical clustering of the RNA-seq analysis results shows differentially expressed genes between dCAR T cells and CAR T cells (n = 2). e Normalised enrichment scores of significantly up- or downregulated gene sets in the CAR T cell versus dCAR T-cell groups (n = 2), as determined by GSEA using the MSigDB C7 gene ontology sets. f Representative GSEA enrichment plot demonstrating the upregulation of memory-related genes and downregulation of differentiation-, exhaustion- and programming death-related genes in dCAR T cells versus CAR T cells (n = 2). g Differentially expressed genes between CAR and dCAR T cells and heatmap demonstrating the different expression profiles of the T-cell function genes for CAR T cells (n = 2). h Mean beta values for the differential CpG sites in the specified genes (n = 2). Significantly differentially CpG sites in (b) and (h) were calculated by generalised linear models (v3.36.2) (P value <0.05, fold change (log2 scale) ≥1 or ≤−1). Significantly differentially expressed genes in (d) and (g) were calculated the Wald test (as implemented in DESeq2) (P value <0.01, fold change (log2 scale) ≥1 or ≤−1).