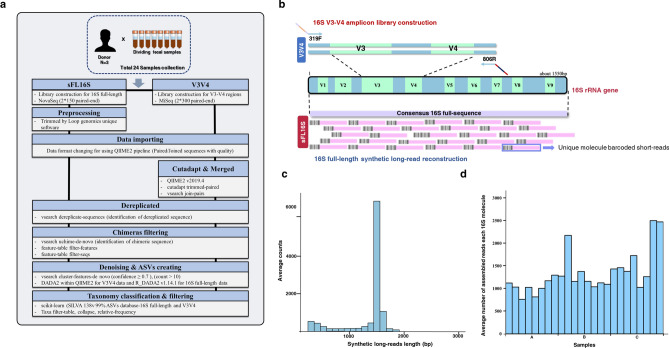
Figure 1.
Technical introduction and analysis workflow for a new 16S full-length-based synthetic long-read (sFL16S) technology. Using 24 fecal samples obtained from three healthy adults, we performed metagenomic analysis and aimed to evaluate an efficient method for microbiome screening and bacterial classification by benchmarking two techniques. (a) The experimental and analytical workflow of two different approaches in the metagenomic analysis of human GUT microbiota. (b) Schematic diagram of the target regions of 16S amplicon library construction for the V3V4 and sFL16S, respectively. For typical 16S amplicon sequencing, the analysis was conducted using the V3V4 variable region. While the short-read targets the V3V4 region, the sFL16S based synthetic long-read targets all variable regions (V1–V9). The high-accurate unique barcoded short-reads (150 PE colored pink), generated from the Illumina Nova-seq were used in the de novo assembly analysis using the UMI tool. (c) Density plot for bp length distribution of synthetic long-read sequencing data. (d) Bar plot for the average number of assembled short-reads each 16S molecule in each sample we used. The X-axis indicates the octuplicate data for each individual.